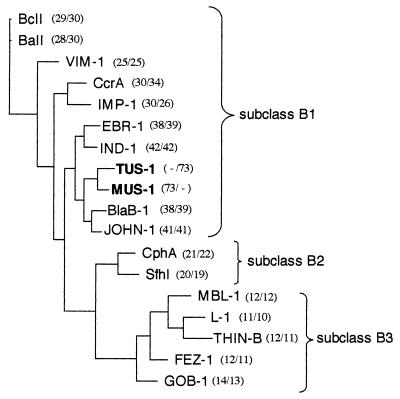
FIG. 3.
Dendrogram obtained by parsimony analysis for representative natural and acquired metallo-β-lactamases. The origins of the metallo-β-lactamases were as follows: BlaB-1 and GOB-1, Chryseobacterium meningosepticum (3); IND-1, Chryseobacterium indologenes (5); EBR-1, Empedobacter brevis (GenBank accession no. AF416700); JOHN-1, Flavobacterium johnsoniae (GenBank accession no. AY028464); CphA, Aeromonas hydrophila (23); SfhI, Serratia fonticola (GenBank accession no. AF197943); L-1, Stenotrophomonas maltophilia (40); BcII, Bacillus cereus (20); BaII, Bacillus anthracis (GenBank accession no. AF367984); VIM-1, Pseudomonas aeruginosa (21); CcrA, Bacteroides fragilis (30); IMP-1, Serratia marcescens (27); THIN-B, Janthinobacterium lividum (31); MBL-1, Caulobacter crescentus (GenBank accession no. AJ315850); and FEZ-1, Legionella gormanii (8). The alignment used for tree calculation was performed with ClustalW followed by minor adjustments. Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The distance along the vertical axis has no significance. Percent amino acid identities to TUS-1 and MUS-1 are indicated in parentheses (left and right numbers, respectively).