Abstract
An epidemic of tuberculosis occurred in a community of Aboriginal Canadians during the period 1987–89. Genetic and epidemiologic data were collected on an extended family from this community, and the evidence for linkage to NRAMP1, a candidate gene for susceptibility to mycobacterial diseases, was assessed. Individuals were grouped into risk (liability) classes based on vaccination, age, previous disease, and tuberculin skin-test results. Under the assumption of a dominant mode of inheritance and a relative risk of 10, which is associated with the high-risk genotypes, a maximum LOD score of 3.81 was observed for linkage between a tuberculosis-susceptibility locus and D2S424, which is located just distal to NRAMP1, in chromosome region 2q35. Significant linkage was also observed between a tuberculosis-susceptibility locus and a haplotype of 10 NRAMP1 intragenic variants. No linkage to the major histocompatibility-complex region on chromosome 6p was observed, despite distortion of transmission from one member of the oldest couple to their affected offspring. The ability to assign individuals to risk classes was crucial to the success of this study.
Introduction
Tuberculosis caused by the human pathogenic bacterium Mycobacterium tuberculosis is a major global public-health problem. In 1997, it was estimated that worldwide there were 7.96 million new cases of tuberculosis (136/100,000 persons), in addition to 8.22 million existing cases, and that 1.87 million people died of tuberculosis (Dye et al. 1999). In Canada, the incidence of tuberculosis is low and was estimated at 7/100,000 (see Appendix 3 in Dye et al. 1999). However, this low rate is not evenly distributed among the Canadian population. Approximately 60% of all cases of tuberculosis occur in immigrants from countries where tuberculosis is endemic, and 15% of the cases occur in the Aboriginal Canadian population, where the annual rate is ∼70/100,000 (Fanning 1999).
It has been argued that people of European origin have a high level of resistance to infection by M. tuberculosis, because of selection for host resistance over 300 years, whereas Aboriginal American populations may be more susceptible, since exposure to M. tuberculosis was rare prior to 1880 (reviewed in Stead 1997). Twin studies (Kallmann and Reisner 1943) and a segregation analysis (Shaw et al. 1997) support a role for a genetic basis of susceptibility. Although there have been many studies examining the relationship between known genes involved in human immune function and tuberculosis, results have not been consistent. A comprehensive summary by Meyer et al. (1998) showed that there is evidence to support a role for the major histocompatibility complex (MHC) region in disease progression but that the role of MHC in resistance to infection is still controversial.
In laboratory strains of inbred mice, it was shown that resistance/susceptibility to the early growth of M. bovis (BCG [Bacille Calmette-Guérin]) is controlled by a locus named “Bcg” and that resistance is inherited as a dominant trait (Gros et al. 1981). Subsequently, on mouse chromosome 1, a gene called “Nramp1” (natural resistance–associated macrophage protein 1) was identified that controls resistance to M. bovis (BCG) and other unrelated intracellular parasites (Vidal et al. 1993, 1995; reviewed in Skamene et al. 1998). The same locus was shown to control early growth of mycobacteria in mice infected with M. lepraemurium, M. intracellulare, M. smegmatis, M. avium, and a group of atypical mycobacteria (reviewed in Schurr and Skamene 1995). The human orthologue, NRAMP1, was cloned and mapped to chromosome region 2q35 (Cellier et al. 1994), and variants within the gene were characterized (Liu et al. 1995a). Associations between four polymorphisms in NRAMP1 and susceptibility to tuberculosis were shown in patients from western Africa; susceptibility appeared to act in a dominant manner, in contrast to the mouse model, where susceptibility is recessive (Bellamy et al. 1998). Susceptibility to leprosy has been linked to NRAMP1 in a sample of 20 nuclear families from Vietnam (Abel et al. 1998). Searle and Blackwell (1999) presented evidence that a polymorphic dinucleotide repeat in the NRAMP1 promoter region influenced gene expression in a luciferase assay. They suggested that an allele that drives low reporter-gene expression contributes to infectious-disease susceptibility, whereas an allele that drives high expression contributes to autoimmune-disease susceptibility. Previously, Blackwell et al. (1997) had reported an association between this promoter-region polymorphism and susceptibility to tuberculosis in 72 age- and sex-matched Brazilian cases and controls. In 37 multicase Brazilian families, Shaw et al. (1997) found weak evidence for linkage between tuberculosis and a highly polymorphic marker adjacent to NRAMP1; however, they found no evidence for linkage to NRAMP1. Hence, the potential role of NRAMP1 in the modulation of tuberculosis susceptibility remains controversial.
An extended Aboriginal Canadian family was exposed to M. tuberculosis and experienced an epidemic of tuberculosis (Mah and Fanning 1991). Given the proved role of Nramp1 in mycobacterial susceptibility in the mouse, human NRAMP1 was chosen as a candidate gene for tuberculosis susceptibility, and linkage analysis was undertaken in this family, primarily to assess the role of NRAMP1.
Subjects and Methods
Family Enrollment and Diagnosis
All subjects enrolled in the present study were members of an extended Aboriginal Canadian family that experienced an outbreak of tuberculosis during 1987–89 (Mah and Fanning 1991). The study was approved by the Research Ethics Board of the University of Alberta. All family members were seen, during the outbreak, by the same physician, and standard forms used by the Alberta Health Tuberculosis Services were completed. Case records and diagnostic criteria were reviewed after the outbreak, and each individual was classified as having a particular disease phenotype (Miller 1991); a synopsis follows. Respiratory-tract specimens (obtained by nasopharyngeal suction or gastric lavage in children) were sent to the Alberta Public Health Laboratory, for mycobacterial culture. A positive culture had either evidence of visible growth on solid media or a growth index >99 in the radiometric system. The species of mycobacteria were identified by conventional methods. In addition, microscopic screening for acid-fast bacilli was done by the auramine-O fluorescent method followed by Kinyoun staining. The disease phenotypes were defined as follows: “culture-positive case”—presence of one or more symptoms or signs associated with tuberculosis (i.e., fever, cough, weight loss, pulmonary infiltrate not responsive to conventional antibiotics, and night sweats) during the period of the epidemic and isolation of M. tuberculosis from respiratory secretions; “culture-negative case”—presence of one or more symptoms or signs associated with tuberculosis, clinical response to antituberculous drugs including chest radiological clearing, and a mycobacteria-negative culture of respiratory secretions; “old case”—tuberculosis disease diagnosed, treated, and presumed to have been cured prior to the epidemic; and “no disease”—absence of disease as defined above. There were no cases of miliary or other forms of extrapulmonary tuberculosis. The culture-negative cases also were negative for acid-fast–bacilli staining; these individuals tended to be young. Chest radiography was done on all subjects. Radiological abnormalities considered to be consistent with active tuberculosis included noncalcified hilar or paratracheal adenopathy and/or parenchymal infiltration. In statistical analyses, we considered culture-positive and culture-negative cases as affected and considered no-disease and old-case individuals as unaffected.
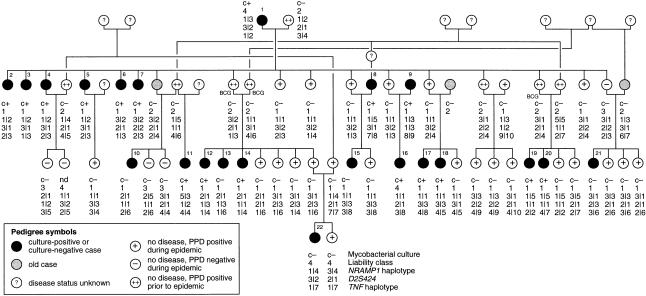
Among previously uninfected individuals who had never been vaccinated with BCG, a new infection with M. tuberculosis was diagnosed by a skin reaction with >10 mm induration, in response to intradermal injection of 5 tuberculin units of purified protein derivative (PPD) (Mah and Fanning 1991). Individuals were assumed to have been previously uninfected if they either (i) had had a prior negative PPD test (14 individuals tested negative during 1968–81, and 15 individuals tested negative early during the epidemic) or (ii) were not known to have had any prior contact with tuberculosis and had had no prior PPD test (24 individuals). A partial pedigree that has been altered for publication is shown in figure 1.
Figure 1 .
Partial pedigree of the extended kindred. The five culture-negative cases (individuals 10, 13, 15, 21, and 22) had radiological abnormalities consistent with active tuberculosis; individuals 10 and 15 also had symptoms associated with tuberculosis. Not included in the pedigree are nine individuals from whom DNA was not available and among whom there were two infants classified as culture-negative cases (both had radiological abnormalities), three children classified as not having disease (all PPD negative; two were culture negative, and no culture was taken from the third), and four people with unknown disease phenotype. The order of the information below each circle is as follows: mycobacterial test result (c+ = M. tuberculosis culture positive; c− = M. tuberculosis culture negative; nd = no data), liability class, NRAMP1 haplotype, D2S424 alleles, and TNF haplotype. D2S424 alleles 1, 2, and 3 are 185, 181, and 157 bp, respectively. BCG indicates that the individual was vaccinated. For classification of disease phenotype and definition of liability classes, see the Subjects and Methods section; for NRAMP1 andTNF haplotypes, see tables 2 and 3, respectively.
Genetic Epidemiology
The risk of developing tuberculosis was assumed to vary among individuals, on the basis of available phenotypic, clinical, and historical information. In order to model gene-environment interaction in a parametric LOD-score linkage model, four risk (liability) classes were created that depended on age, BCG-vaccination status, tuberculin skin-test results, and previous infection and disease. Each individual was assigned to one liability class. The penetrances in each class were estimated on the basis of published studies and reviews of tuberculosis, as well as on the basis of observations of this extended family and the surrounding community (Boothroyd 1994); table 1 gives the assumed penetrances for the four liability classes. Liability class 1 consisted of individuals who, as the result of exposure to tuberculosis during the epidemic, converted from PPD negative (i.e., either known to be negative or, in the absence of test results, assumed to be negative) to PPD positive. The penetrance of the high-risk genotype was assumed to be .85. Approximately 15% of the family members tested PPD negative during the epidemic, and therefore this number was used as an index of insufficient exposure. Liability class 2 included individuals who either (a) previously had been vaccinated with BCG, (b) were PPD positive prior to the epidemic but were not known to have had the disease, or (c) previously had been affected with tuberculosis and had been successfully treated. These individuals were assumed to have a lower risk of disease at subsequent exposure (Ferguson 1955). The penetrance of the high-risk genotype (i.e., .37) was based on a case-control study of vaccine efficacy in Aboriginal Canadians that showed a 57% reduction in risk (Houston et al. 1990). Liability class 3 included individuals who were PPD negative for tests at two different times and who therefore may have been unexposed or not intensively exposed to M. tuberculosis. Their high-risk genotype penetrance of .10 was adapted from an estimate of the false-negative rate in PPD tests (Holden et al. 1971; Boothroyd 1994). Finally, liability class 4 included individuals who were ⩽2 or ⩾65 years of age and, because of an immature or a compromised immune system, were considered to be at high risk of disease. Their high-risk penetrance was assumed to be .85, as in liability class 1. The genotype relative risk, or the ratio of the penetrance of the high-risk genotype to the low-risk genotype, was assumed to be 10 in liability classes 1–3, but in liability class 4, even in individuals who did not carry any susceptibility alleles, the risk of disease was assumed to be much higher, such that a relative risk of 2 was assumed for this group. The frequency of the high-risk allele was assumed to be .20 for a recessive model and .05 for a dominant model; a fairly high allele frequency is plausible, given the history of the community. Estimated disease prevalences were based on the dominant-model parameters (table 1). The resulting estimated prevalences are high (e.g., 16% in liability class 1) when compared with published tuberculosis incidence rates, but there is evidence to support a very high susceptibility among Aboriginal Canadian peoples (Ferguson 1955).
Table 1.
Penetrance Values and Prevalences for Linkage Analysis, for the Phenotype of Tuberculosis Disease
|
Penetrance ofb |
No. of Individuals |
||||
| Liability Classa | Low-Risk Genotype | High-Risk Genotype | Estimated Disease Prevalencec | Total | Affected |
| 1 | .085 | .85 | .16 | 42 | 19 |
| 2 | .037 | .37 | .07 | 11 | 0 |
| 3 | .010 | .10 | .02 | 7 | 0 |
| 4 | .425 | .85 | .47 | 7 | 5 |
| Unknownd | 14 | … | |||
1 = Previously unexposed; 2 = previously exposed, vaccinated, or previously treated for disease; 3 = PPD negative during epidemic; 4 = very young or elderly.
Penetrance of the heterozygote was assumed to be equal to that of either the low-risk or the high-risk homozygote, depending on whether a recessive or a dominant mode of inheritance, respectively, was assumed.
Based on an assumed disease-allele frequency of .05, a dominant mode of inheritance, and the penetrances shown.
Ancestors not exposed during the epidemic.
Twenty-nine markers on chromosome 2q, spanning ∼71 cM, were typed, including 10 variants in either NRAMP1 or the immediate flanking regions of the gene (fig. 2). NRAMP1 haplotypes were deduced from the genotypes by inspection of their segregation in the pedigree; table 2 shows the haplotype codes. Only one polymorphism in this study corresponds to a missense variant (D543N), and analysis of this diallelic polymorphism was undertaken separately. A separate analysis was also undertaken for the flanking region 5′ (GT)n dinucleotide repeat (Liu et al. 1995a); allele 286 corresponds to allele 3 in the study by Searle and Blackwell (1999), which was shown by these investigators to drive high gene expression in vitro. In addition, on chromosome 6, two single-nucleotide polymorphisms (SNPs) in the promoter region of the tumor necrosis–factor gene (TNF) and six nearby markers were genotyped, and segregating haplotypes were inferred (table 3).
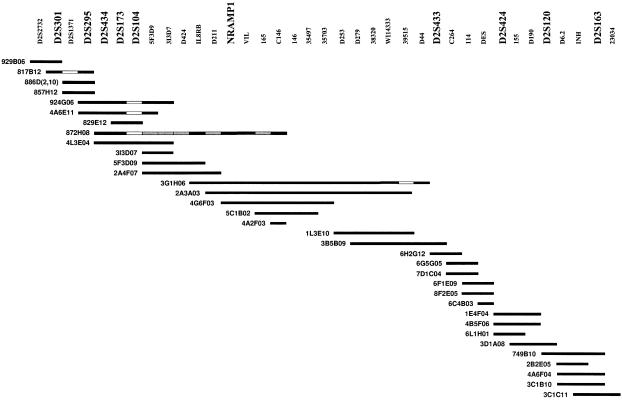
Figure 2.
YAC contig of the NRAMP1-gene region. The names of loci and chromosome 2–specific IRS probes are indicated at the top of the figure. YAC clones are represented as bars; the YAC address is given to the left of the bar. The blackened area of each bar indicates that the YAC was unambiguously identified by the probe listed directly above the YAC; the unblackened area of each bar indicates that the probe did not hybridize with the YAC; and the gray-shaded area of each bar indicates that the YAC was not tested for the presence of the probe. D2S1471 was not included in STS contig mapping; the order NRAMP1–VIL–D2S1471 was established by physical mapping in a PAC contig (Abel et al. 1998).
Table 2.
NRAMP1 Haplotypes
|
Haplotype |
||||||
| Marker | Location in NRAMP1 | 1 | 2 | 3 | 4 | 5 |
| (GT)na,b,c | 5′ Promoter region | 286 | 286 | 288 | 288 | 286 |
| 77−385C/Td | 5′ Promoter region | C | T | C | C | C |
| 274C/Ta | Codon 66, exon 3 | C | C | T | C | C |
| 469+14G/Ca,c | Intron 4 | G | G | C | G | G |
| 577−18G/Aa | Intron 5 | G | G | G | G | G |
| 823C/Ta | Codon 249, exon 8 | C | C | C | T | T |
| A318Va | Codon 318, exon 9 | Ala | Ala | Ala | Ala | Ala |
| 1465−85G/Aa | Intron 13 | G | G | A | A | A |
| D543Na,c | Codon 543, exon 15 | Asp | Asn | Asp | Asn | Asn |
| (CAAA)ne | 3′ UTR | 2 | 3 | 3 | 3 | 3 |
Name of variant as defined by Liu et al. (1995a); the nucleotides and codons were numbered according to the sequence of GenBank accession number L32185.
Microsatellite alleles are designated by size (in bp), SNPs by nucleotides, coding variants by amino acids, and the insertion/deletion polymorphism by number of repeat units.
Marker used in the case-control study of western Africans, by Bellamy et al. (1998).
Table 3.
Haplotypes in the TNF Region
|
Haplotype |
||||||||||||
| Markera | Type | Location b | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| TNFb | (GA)n | 5,119 bp 5′ to initiation codon of LTA | 131 | 130 | 131 | 131 | 129 | 130 | 127 | 131 | 131 | 131 |
| TNFa | (GT)n | 5,066 bp 5′ to initiation codon of LTA | 97 | 109 | 109 | 107 | 99 | 115 | 99 | 107 | 103 | 103 |
| TNFn | SNP | Intron 1 of LTA | T | T | C | C | C | T | T | C | C | T |
| TNFc | (GA)n | Intron 1 of LTA | 161 | 159 | 159 | 159 | 159 | 159 | 161 | 159 | 159 | 161 |
| TNFα-308 | SNP | Promoter region of TNF | C | C | C | C | T | C | C | C | T | C |
| TNFA-238 | SNP | Promoter region of TNF | T | C | C | C | C | C | C | C | C | C |
| TNFe | (TC)n | Intron 4 of LST1 | 104 | 104 | 104 | 104 | 104 | 104 | 100 | 104 | 104 | 104 |
| TNFd | (TC)n | Intron 4 of LST1 | 130 | 130 | 130 | 134 | 126 | 130 | 132 | 130 | 130 | 134 |
Genotyping
DNA samples were genotyped for microsatellite markers (see table 4) that flank NRAMP1 in chromosome region 2q35, by standard radioactive protocols (Hudson et al. 1997) with minor modifications. The conditions for genotyping 8 of the 10 intragenic NRAMP1 markers have been described elsewhere (Liu et al. 1995a). The insertion/deletion polymorphism (CAAA)n was amplified by the forward primer reported by Buu et al. (1995) and by the reverse primer 5′-TCAAGCTCCAGTTTGGAGCCT-3′ and, by electrophoresis on 10% polyacrylamide gels stained with ethidium bromide, was genotyped as a length variant. The SNP, 77−385C/T, identified by Lewis et al. (1996) was genotyped by primers 5′-GGGTGTGGTCATGGGGTATTGA-3′ and 5′-CAAAGGCAGAAGTGGCCAG-3′, in a similar manner to that used for other NRAMP1 SNPs (Liu et al. 1995a). The allele with nucleotide T creates an Msl I restriction site. Digestion of the 202-bp PCR fragment by Msl I results in 175-bp and 27-bp fragments for allele T and in the 202- bp fragment for allele A. In the MHC region on chromosome 6, we genotyped five microsatellite markers (TNFa, TNFb, TNFc, TNFd, and TNFe) as described by Udalova et al. 1993, and three SNPs referred to here as “TNFn” (Udalova et al. 1993), “TNFα-308” (Wilson et al. 1992), and “TNFA-238” (D'Alfonso and Richiardi 1994). SNPs were genotyped by assays based on naturally occurring or amplification-created restriction sites. With minor modifications, TNFn was genotyped as described by Roth et al. (1994), TNFα-308 was genotyped as described by Wilson et al. (1992), and TNFA-238 was genotyped by Ava II digestion of fragments amplified by primer 5′-CAAACACAGGCCTCAGGACTC-3′ (McGuire et al. 1994) and a primer—5′-TCACACTCCCCATCCTCCCTGGTC-3′—modified to create an Ava II site with one of the alleles.
Table 4.
LOD Scores for Linkage under a Dominant Model, Genotype Relative Risk 10, and Disease-Allele Frequency .05
|
LOD Score at Recombination Fraction of |
|||||||
| Marker | Composite Locationa(Mb) | .00 | .01 | .05 | .10 | .20 | .40 |
| Chromosome 2: | |||||||
| HOXD8 | 181.66 | −2.91 | −2.71 | −2.06 | −1.48 | −.76 | −.17 |
| D2S152 | 194.26 | −1.65 | −1.58 | −1.29 | −.98 | −.50 | −.05 |
| CRYG | 221.82 | 1.22 | 1.21 | 1.13 | 1.00 | .66 | .07 |
| D2S157 | 222.80 | 1.38 | 1.42 | 1.49 | 1.45 | 1.12 | .08 |
| D2S128b | 226.87 | 1.23 | 1.28 | 1.37 | 1.37 | 1.12 | .12 |
| D2S137b | 228.80 | 2.17 | 2.11 | 1.88 | 1.59 | 1.03 | .10 |
| TNP1 | 220.26 | 1.49 | 1.47 | 1.34 | 1.16 | .78 | .16 |
| D2S301 | 230.49 | 2.08 | 2.06 | 1.95 | 1.75 | 1.26 | .22 |
| D2S164 | 230.60 | −.51 | −.51 | −.48 | −.43 | −.28 | −.01 |
| D2S434 | 233.15 | 2.55 | 2.52 | 2.39 | 2.18 | 1.62 | .19 |
| D2S173c | 232.32 | 2.71 | 2.72 | 2.65 | 2.45 | 1.86 | .23 |
| D2S104c | 232.96 | −.90 | −.88 | −.80 | −.71 | −.47 | −.06 |
| NRAMP1 (GT)nd | 227.21 | 3.55 | 3.49 | 3.26 | 2.92 | 2.16 | .32 |
| NRAMP1 D543Nd | 227.21 | 3.21 | 3.18 | 2.99 | 2.71 | 2.02 | .30 |
| NRAMP1 haplotype | 227.21 | 3.36 | 3.32 | 3.16 | 2.90 | 2.23 | .38 |
| D2S1471 | … | .79 | .79 | .78 | .75 | .61 | .14 |
| D2S433 | 232.84 | .47 | .49 | .52 | .51 | .38 | −.07 |
| D2S424 | 235.92 | 3.81 | 3.77 | 3.57 | 3.24 | 2.40 | .35 |
| D2S102 | 236.93 | −.51 | −.50 | −.45 | −.40 | −.32 | −.10 |
| D2S126 | 236.26 | 3.11 | 3.09 | 2.96 | 2.71 | 2.04 | .28 |
| D2S172 | 246.68 | 1.20 | 1.25 | 1.37 | 1.39 | 1.16 | .10 |
| D2S125 | 253.37 | −2.93 | −2.82 | −2.41 | −1.90 | −1.04 | −.16 |
| Chromosome 6: | |||||||
| TNFa | … | −.07 | −.07 | −.05 | −.04 | −.02 | .00 |
| TNFb | 37.81 | −1.48 | −1.39 | −1.07 | −.78 | −.42 | −.12 |
| TNF haplotype | 37.81 | −2.64 | −2.44 | −1.79 | −1.18 | −.48 | −.10 |
From the Genetic Location Database, updated November 9, 1999. The order of the markers shown, from TNP1 to D2S126, is based on physical mapping data, not on the Genetic Location Database estimated order.
The physical order of the markers within the pair D2S128 and D2S137 was not resolved.
The physical order of the markers within the pair D2S173 and D2S104 was not resolved.
The NRAMP1 orientation with respect to flanking markers has been derived from genomic sequence information (S. Marquet and E. Schurr, unpublished data).
Physical Mapping
To establish the order of microsatellites in the immediate vicinity of NRAMP1, we established a YAC contig spanning an estimated 2.5×106 bp of genomic DNA in the NRAMP1 region. The YAC libraries screened consisted of the Centre d'Étude du Polymorphisme Humain (CEPH) Mark I YAC library plates numbered 1–552, plus 24 plates of the original Washington University YAC library. This setup of library plates has been described elsewhere (Liu et al. 1995b). In addition, the CEPH Mega YAC library plates 1–860 and 869–972 were used. From all YAC clones, interspersed repetitive sequences (IRS)–PCR products were prepared and spotted on nylon membranes, as described elsewhere (Liu et al. 1995b). The filters were screened by chromosome 2–specific IRS probes, and extended chromosome 2 YAC contigs were obtained (Liu 1996). Microsatellites (D2S301, D2S1371, D2S295, D2S434, D2S173, D2S104, D2S433, D2S424, D2S120, and D2S163) were selected from published genetic maps and were placed on the YAC contig map overlapping the NRAMP1 region. In addition, we selected primer pairs for five known genes (IL8RB, NRAMP1, VIL, DES, and INHA) and one sequence-tagged site (STS) (WI14333). The source, sequence, and conditions for amplification, except for WI14333, have been described elsewhere (Liu et al. 1995b). The sequence and conditions for amplification of WI14333 were taken from the Whitehead Institute for Biomedical Research/MIT Center for Genome Research Web site. All contigs were subjected to STS contig mapping, and several contigs in chromosome region 2q35, which contain NRAMP1, were identified. These contigs were integrated with the Whitehead Institute for Biomedical Research/MIT Center for Genome Research YAC map, and the remaining contig gaps in the NRAMP1 region were closed by chromosome walking in YAC libraries (Liu et al. 1995b; Liu 1996). The relative position of microsatellite marker D2S1471 was determined by physical mapping in a plasmid artificial chromosome (PAC) contig, as described elsewhere (Abel et al. 1998).
Statistical Methods
Two-point LOD scores for the four liability-class gene-environment–interaction model (described above) were calculated by FASTLINK version (4.1P) (Cottingham et al.1993; Schäffer et al. 1994) of the LINKAGE programs (Lathrop and Lalouel 1984; Lathrop et al. 1984, 1986). The pedigree contained four loops. Marker-allele frequencies in the founders of the pedigree were estimated by the ILINK program (Lathrop et al. 1984), under the assumption of linkage equilibrium with each marker unlinked to a dummy locus (Terwilliger and Ott 1994). Model-based linkage analysis was performed under both recessive and dominant modes of inheritance (Hodge et al. 1997; Morton 1998; Durner et al. 1999), and the genotype relative risk of disease was varied from 10 to 100, for liability classes 1–3, and from 2 to 4, for liability class 4. Since the diagnosis of tuberculosis was made in some cases by clinical information rather than by positive cultures, the sensitivity of the results to each diagnosis was assessed. One by one, the phenotype of each affected individual was altered to “unknown,” and the analysis under a dominant model was repeated. Nonparametric linkage analysis (Kruglyak 1997) based on affected-member allele sharing (e.g., see Kruglyak et al. 1996) was not attempted, because of both the size of the pedigree and the presence of loops; however, a model-free analysis was performed by a modified version of the MFLINK program (Curtis and Sham 1995; C. M. T. Greenwood and K. Morgan, unpublished data), a version that allows more than one liability class.
Diallelic polymorphisms in NRAMP1 were tested for association with disease, by examination of transmissions from heterozygous parents. Since all transmissions occur within the same family, under the assumption of linkage without association the transmissions are not independent. In fact, most of the counted transmissions are due to the segregation of one allele from the oldest couple to their children and grandchildren. Therefore, it was not possible to assess whether, in addition to linkage, there is significant association with particular alleles.
Results
The epidemic in this rural community was carefully documented by Mah and Fanning (1991). One member returned to live with the family and then developed symptomatic tuberculosis that was undiagnosed for 4–5 mo. Contact between all family members was thought to be frequent; all nuclear families lived close to one another. After diagnosis of the index case, a screening program was implemented, and individuals were treated as necessary. The extended family of 81 individuals (fig. 1) includes 67 individuals who were clinically assessed, of whom 61 were genotyped. An additional 14 ancestors were included, to link members of the pedigree. Forty-two individuals tested as being newly PPD positive during the epidemic and, prior to the epidemic, had not been either vaccinated or diagnosed as having tuberculosis. These individuals were assigned to liability class 1 (table 1). Liability class 2 included 11 individuals: 3 individuals who previously had been vaccinated with BCG, 5 who were PPD positive prior to the epidemic, and 3 who previously had been affected with tuberculosis and had been successfully treated. Seven individuals who never tested as PPD positive were assigned to liability class 3, and seven very young or elderly individuals were assigned to the high-risk class 4. Of the 67 observed individuals, all 42 in liability class 1 were genotyped, 10 of 11 in class 2, 4 of 7 in class 3, and 5 of 7 in class 4.
Twenty-four members of the family became affected with active tuberculosis during the 2 years following the diagnosis of the index case (table 1). Seven individuals were culture negative but were diagnosed by symptoms, confirmatory X-ray, and response to treatment. All seven were <10 years of age (5 of these 7 were <5 years old); the sensitivity of culture diagnosis in young children is known to be poor (Smith and Marquis 1987). Four individuals had been diagnosed with tuberculosis >10 years prior to this epidemic. One of these old cases (individual 1; see fig. 1) developed active disease during the outbreak; this case may have been due to reactivation of old disease, rather than to a new infection. All cases were appropriately treated; in addition, control measures led to izoniazid treatment for most PPD-positive individuals and other individuals identified as being at high risk (Mah and Fanning 1991).
NRAMP1 Region
Table 4 shows the results of a LOD-score analysis under a dominant model using the penetrances in table 1. For the purposes of this analysis, the individual who experienced a second case of tuberculosis was considered to be newly affected. The maximum LOD score of 3.81 was obtained for a recombination fraction of 0, with D2S424, which is estimated to be 8.71 Mb away from NRAMP1 (Genetic Location Database). Physical-map data, however, estimate the distance to be <2×106 bp. Under a recessive model, the maximum LOD score for D2S424 was 2.8 and again occurred at 0 recombination. Informativity of many markers in the region was incomplete. The NRAMP1 haplotype contributes 77 equivalent (informative) meioses (Ott 1991), of a possible 97, whereas the NRAMP1 missense variant (D543N; also included in the NRAMP1 haplotype) contributed 40 informative meioses.
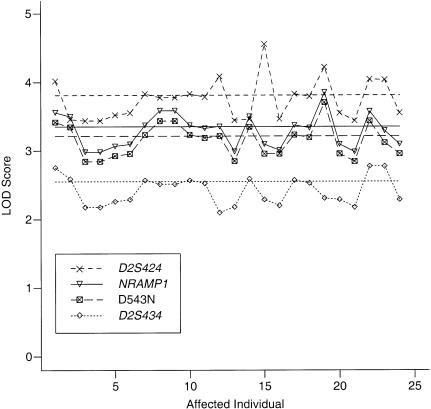
Figure 3 illustrates the sensitivity of the results to the disease status. One by one, the status of each affected individual was altered to be unknown. The first point on the left corresponds to the individual in the family whose illness may have been due to a reactivation of previous disease. The LOD score rose by ∼0.2 when the affection status of this individual was considered to be unknown instead of affected. Despite the variability, none of the LOD scores for the NRAMP1 haplotype fell to <3, and none of the LOD scores for D2S424 fell to <3.5. When all high-risk individuals together in class 4 were assigned an unknown phenotype, the LOD scores changed very little from their original values. Minimal changes also were seen when the seven individuals in liability class 3, who may have had false-negative PPD tests, all were assigned an unknown affection status.
Figure 3 .
Sensitivity of LOD-score results to the diagnosis of an affected individual, for the NRAMP1 haplotype, two flanking microsatellite markers, and one intragenic polymorphism, D543N. LOD scores in all cases were maximized over the recombination fraction. Horizontal lines show the LOD scores for the original data. The order along the horizontal axis corresponds to the identification numbers of affected individuals in figure 1, with the exception of individuals 23 and 24, who were two culture-negative cases not shown in figure 1.
Results varied slightly when the disease-allele frequency or the genotype relative risk were changed. The frequency of allele Asn at codon 543 of NRAMP1 was estimated to be 21% in the founders of the pedigree; however, this allele is rare in Canadians of European ancestry (Liu et al. 1995a). Reducing the allele frequency to 1% in the linkage analysis resulted in a small increase in the maximum LOD score for this marker, from 3.31 to 3.41. In order to evaluate the dependence of the results on the assumed genetic model in a more robust manner, the model-free approach of Curtis and Sham (1995) was altered to allow multiple liability classes (C. M. T. Greenwood and K. Morgan, unpublished data). Assuming that the susceptibility locus could not confer a genotype risk >10, the algorithm searched over allele frequencies and penetrances for dominant and recessive models that were consistent with the prevalences in table 1. A model-free LOD score of 2.68 was obtained for the NRAMP1 haplotype.
Because of the size and complexity of the pedigree, multipoint analysis using all markers was impractical. However, linkage analysis was undertaken with two markers jointly (the NRAMP1 haplotype and D2S424), and a maximum LOD score of 4.2 was obtained between the two marker loci. Similarly, three-point linkage-analysis models that paired the NRAMP1 haplotype with either D2S433, D2S1471, or D2S434 gave multilocus LOD scores of 3.1, 3.8, and 3.8, respectively, and each analysis estimated the location of the susceptibility locus to be either at the NRAMP1 haplotype or between the second marker and the NRAMP1 haplotype.
The alleles at all marker loci in the region were examined for association with disease status. Allele Asn of the diallelic missense variant, D543N, appeared to be associated with disease. In transmissions to affected children from a heterozygous parent (when both parents were typed), there were 11 transmissions of allele Asn, versus only 2 transmissions of allele Asp; however, this apparent association is mostly attributable to the linkage of disease with the NRAMP1 haplotype 2. Similarly, at 77−385C/T, which is in the promoter region of NRAMP1, allele T was transmitted 10 times (vs. 0 transmissions of allele C) from heterozygous parents. It can be seen in figure 1 and table 2 that this pattern is entirely due to the segregation of the NRAMP1 haplotype 2 from individual 2. At the promoter-region polymorphism (GT)n, we observed four transmissions of allele 288, versus 13 transmissions of allele 286, from heterozygous parents.
HLA Region
Although the HLA region is thought to have a role in tuberculosis progression (Meyer et al. 1998), it is still unresolved whether genes in this region contribute to tuberculosis susceptibility. In this pedigree, there was no evidence for linkage between the susceptibility locus and markers either in or near TNF (table 4) that are in the HLA class III region; however, the six affected offspring of the oldest couple all inherited identical TNF and NRAMP1 haplotypes from one of their parents (table 5). This pattern of cosegregation of two unlinked loci was unexpected, and potential reasons for the joint transmissions were explored.
Table 5.
Segregation of NRAMP1 and TNF Haplotypes from the Spouse of Individual 1 to 17 Offspring
|
Haplotypea |
|||
| Status | NRAMP1 | TNF | No. of Siblings (Liability Class) |
| Affected b | 2 | 3 | 6 (LC 1) |
| Unaffected | 2 | 3 | 1 (LC 2) |
| 1 | 3 | 1 (LC 3), 3 (LC 1) | |
| 2 | 4 | 1 (LC 2) | |
| 1 | 4 | 3 (LC 1), 2 (LC 2) | |
All affected offspring received the same haplotypes at both genes. One BCG-vaccinated unaffected individual also received this haplotype combination, but all other unaffected individuals received different haplotype combinations.
The possibility of gene-gene interaction between TNF and NRAMP1 was evaluated in several ways. We did not fit a two-gene model, because of the complexity of the pedigree. However, we evaluated linkage to the TNF haplotype while allowing the penetrances to vary with the allele carried at D543N in NRAMP1. Similarly, we evaluated linkage to the NRAMP1 haplotype and allowed the penetrances to vary between carriers of the TNF haplotype transmitted to the six affected offspring of the second generation (haplotype 3 in table 3) and carriers of all other alleles. LOD scores for linkage to the TNF haplotype were consistently <0, under several interaction models. Because of insufficient data, we were unable to estimate the penetrances in these models with eight liability classes.
The pattern of joint transmission to the affected siblings of the second generation might also be due to segregation distortion that operates jointly on TNF and NRAMP1. We examined the transmission of TNF and NRAMP1 haplotypes for each typed parent-child pair in the pedigree and found neither evidence for biased segregation overall nor statistical evidence for a segregation-pattern difference due to the gender of the parent who transmitted the alleles (P=.5).
If it is assumed that near to NRAMP1 there is a susceptibility locus for tuberculosis but that the MHC locus does not affect disease susceptibility, this pattern of joint transmission could have occurred by chance. The probability that this would occur by chance can be calculated as .037 (by examination of the probability of transmissions of the TNF haplotype to a fixed set of 6 affected individuals and to 5 of 11 unaffected individuals, given that there were 11 transmissions to 17 children). If there were no susceptibility locus near NRAMP1, then the probability that the six affected siblings would receive the alleles at two loci would be much smaller than 4%.
Discussion
An extended Aboriginal Canadian family that experienced an epidemic of tuberculosis has shown evidence for linkage between susceptibility for symptomatic disease and chromosome 2q35 loci near a strong candidate gene, NRAMP1. The model-based analysis used four liability classes in which the chosen high-risk penetrances were based on the epidemiology of the disease. It has been shown that, when the true mode of inheritance is unknown, linkage can be robustly detected by estimation of LOD scores for a small number of different sets of penetrances (incomplete penetrance, dominant and recessive models, and, perhaps, a range of genotype relative risks) and that this approach will lead to only a small increase in the type I error rate (Hodge and Elston 1994; Hodge et al. 1997; Morton 1998; Durner et al. 1999).
The choice of penetrances (table 1), although, as far as possible, based on knowledge of the disease epidemiology, was somewhat arbitrary. Hence, we developed and used a model-free approach, with multiple liability classes, that maximized separately the numerator and the denominator of the linkage likelihood ratio over the disease-allele frequency, the mode of inheritance, and penetrances that were compatible with fixed disease prevalences. This robust analysis also found substantial evidence in favor of linkage and lends support to the traditional analysis.
In two-point linkage analysis, the estimated recombination fractions between the susceptibility locus and marker loci were all nearly 0 for markers close to or within NRAMP1. In analyses that included two markers and the susceptibility locus, the location scores also estimated that the susceptibility locus was very near to NRAMP1. However, it is known that multipoint analysis can result in more-biased estimates of the disease-gene location if other parameters of the model are misspecified (Risch and Giuffra 1992; Terwilliger and Ott 1994; Halpern and Whittemore 1999). In all cases, a 1-LOD support interval for the location of the disease gene spanned 10–15 cM on each side of NRAMP1. In fact, for multilocus models, Terwilliger and Ott (1994) recommended use of an interval wider than 1 LOD. Therefore, we cannot rule out the possibility that tuberculosis susceptibility is affected by a gene closely linked to but distinct from NRAMP1.
The construction of four liability classes defining different individual risks of overt disease was critical to obtaining significant evidence for linkage. Although, for complex diseases, it has been shown that simple LOD-score models can outperform nonparametric methods when the true model of inheritance is unknown (Hodge et al. 1997; Greenberg et al. 1998; Abreu et al. 1999), the availability of additional information about risk categories will improve the power to detect linkage (Hodge et al. 1980). For an infectious disease such as tuberculosis, sufficient exposure to the causative agent is critical, and the incidence rate will increase with exposure intensity (Ferguson 1955). Unfortunately, it is not possible to accurately measure exposure intensity. The four liability classes represent our best assessments of both the exposure and the susceptibility of each individual. An analysis that used these classes found strong evidence for linkage for a variety of assumed penetrances and disease-allele frequencies. However, an analysis that did not make use of this epidemiological information did not find significant evidence for linkage, no matter what parameter values were assumed (data not shown).
Although allele 286 of the dinucleotide repeat in the promoter region of NRAMP1 was reported to have higher gene expression in vitro and was postulated to be protective against infectious disease (Searle and Blackwell 1999), we observed excess transmission of this allele from heterozygous parents to affected children. However, this finding does not provide any evidence for or against a disease association with this allele, since association and linkage are confounded in these transmissions.
An attempt was made to study a second epidemic in a similar setting, for the purposes of validating the observed linkage to NRAMP1. During 1994–95, an epidemic of tuberculosis occurred in another community of Aboriginal Canadians, who were from the same cultural and linguistic heritage. A pedigree of 101 individuals was analyzed for linkage; 18 individuals were diagnosed as having tuberculosis. There was no evidence for or against linkage, since LOD scores were close to 0 (data not shown). There was a higher proportion of individuals who were PPD negative, and the proportion of individuals who contributed blood samples was lower than that in the primary epidemic. Geographic and social contact between family members was not as close. Furthermore, because of both the number of individuals not genotyped and the pattern of affected individuals observed, the potential power to detect linkage was low. No other epidemic of tuberculosis in Aboriginal Canadians from the same cultural and linguistic heritage has become available for study.
In the mouse, Nramp1 does not appear to affect susceptibility to M. tuberculosis (Medina and North 1996; North et al. 1999). The strongest evidence comes from the North et al. (1999) study that used one strain of M. tuberculosis (H37Rv) and 129sv mice homozygous for functional deletion of Nramp1. Since M. tuberculosis is not a natural pathogen of rodents, and since, in several important aspects, M. tuberculosis–induced pathology in mice differs from the human disease (e.g., in mice, there is no latent phase, and the extent of granuloma formation is different than that in humans), it is debatable whether infection of laboratory mice with M. tuberculosis represents a good model for human tuberculosis. By contrast, M. microtis, a member of the M. tuberculosis complex, is a natural pathogen of field and forest mice and causes a tuberculosis-like disease (Liébana et al. 1996). Systematic investigations that address the genetic control of M. microtis infection in laboratory mice are not available at present.
Because of the nature of the contacts between family members in this kindred, most individuals probably were exposed intensely to M. tuberculosis early during the epidemic and became infected (table 1). Therefore, the linkage of tuberculosis to NRAMP1 is consistent with a phenotype of progression to overt disease rather than susceptibility to infection, although, in the mouse, Nramp1 controls early mycobacterial growth (Schurr and Skamene 1995). Furthermore, mouse Nramp1 acts dominantly to confer early-phase resistance to mycobacterial infection, whereas a dominant model for susceptibility appeared to be more likely in this family and in the western Africans studied by Bellamy et al. (1998). Allelic variants in mice differ in controlling the growth rate of intracellular parasites (Vidal et al. 1995); both intra- and interspecies differences in allelic variants and in their function should be expected.
NRAMP1 appears to play a substantial role in affecting disease susceptibility in this family. Under the genetic model assumed here (table 1), carriers, in liability classes 1–3, of at least one copy of a high-risk allele have a 10-fold-increased risk of disease, compared with individuals with no copies of the high-risk allele. However, in the study by Bellamy et al. (1998), NRAMP1 variants were estimated as conferring small relative risks for tuberculosis susceptibility (odds ratios <2.0), and other studies have failed to find linkage of tuberculosis to NRAMP1 (Shaw et al. 1997). In The Gambia, almost all individuals had been infected (Bellamy et al. 1998). In the Canadian community, BCG vaccination lapsed in 1963 (Mah and Fanning 1991), and only a few individuals had been exposed to tuberculosis prior to the epidemic. Hence, family members likely had low levels of specific anti–M. tuberculosis immunity, and almost everyone might be expected to have been susceptible to infection and disease. Furthermore, between the arrival of the index case in the community and the initiation of a general treatment and control program that would have lowered the exposure intensity, there was a period of intense exposure lasting several months. Thus, in this family, NRAMP1 appears to modulate the rate at which individuals progress from being infected (i.e., PPD positive) to having symptomatic disease. Results may have been very different if (a) the study had been undertaken either in a context where exposure was less intense, perhaps as in the attempted validation sample, or under pandemic rather than epidemic conditions where most individuals are continuously exposed to M. tuberculosis or (b) if the timing of the treatment and control program had been different. In conclusion, in the epidemiological context of a tuberculosis outbreak in a community of Aboriginal Canadians, most of whom had not been BCG vaccinated, there is significant evidence to support a role for NRAMP1 or a gene (or genes) closely linked to NRAMP1 in susceptibility to active tuberculosis disease.
Acknowledgments
We are grateful to all the individuals who participated in our study. We thank Ann Josée Paradis for genotyping and laboratory expertise, Dianne Binns for collection of samples and clinical data, Jing Liu and Jiang-Xue Wang for genotyping and physical mapping, Joyce Crumley for database management, Silvia Vidal for helpful discussions, and two anonymous reviewers for their helpful comments. This work was supported by grants from the Networks of Centres of Excellence program—the Canadian Genetic Diseases Network and the Mathematics of Information Technology and Complex Systems—and by Pasteur Mérieux Connaught Canada. C.M.T.G. was supported by a postdoctoral fellowship from the Medical Research Council of Canada, T.M.F. by a gift to McGill University from Alcan Aluminum Limitée, and E.S. by a career award from the Fonds de la recherche en santé du Québec
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for nucleotide positions [accession numbers D63507, L32185, U57604, and Y14768])
- Genetic Location Database, The, http://cedar.genetics.soton.ac.uk/public_html/ldb.html (for genetic distances and locations)
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www-genome.wi.mit.edu (for sequence and conditions of amplification of WI14333)
References
- Abel L, Sánchez FO, Oberti J, Thuc NV, Hoa LV, Lap VD, Skamene E, et al (1998) Susceptibility to leprosy is linked to the human NRAMP1 gene. J Infect Dis 177:133–145 [DOI] [PubMed] [Google Scholar]
- Abreu PC, Greenberg DA, Hodge SE (1999) Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. Am J Hum Genet 65:847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy R, Ruwende C, Corrah T, McAdam KPWJ, Whittle HC, Hill AVS (1998) Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med 338:640–644 [DOI] [PubMed] [Google Scholar]
- Blackwell JM, Black GF, Peacock CS, Miller EN, Sibthorpe D, Gnananandha D, Shaw JJ, et al (1997) Immunogenetics of leishmanial and mycobacterial infections: the Belem Family Study. Philos Trans R Soc Lond B Biol Sci 352:1331–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothroyd LJ (1994) Genetic susceptibility to tuberculosis. MSc thesis, Department of Epidemiology and Biostatistics, McGill University, Montreal [Google Scholar]
- Buu NT, Cellier M, Gros P, Schurr E (1995) Identification of a highly polymorphic length variant in the 3′UTR of NRAMP1. Immunogenetics 42:428–429 [DOI] [PubMed] [Google Scholar]
- Cellier M, Govoni G, Vidal S, Kwan T, Groulx N, Liu J, Sanchez F, et al (1994) Human natural resistance-associated macrophage protein: cDNA cloning, chromosomal mapping, genomic organization and tissue-specific expression. J Exp Med 180:1741–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Sham PC (1995) Model-free linkage analysis using likelihoods. Am J Hum Genet 57:703–716 [PMC free article] [PubMed] [Google Scholar]
- D'Alfonso S, Richiardi PM (1994) A polymorphic variation in a putative regulation box of the TNFA promoter region. Immunogenetics 39:150–154 [DOI] [PubMed] [Google Scholar]
- Durner M, Vieland VJ, Greenberg DA (1999) Further evidence for the increased power of LOD scores compared with nonparametric methods. Am J Hum Genet 64:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC for the WHO Global Surveillance and Monitoring Project (1999) Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. JAMA 282:677–686 [DOI] [PubMed] [Google Scholar]
- Fanning A (1999) Tuberculosis. I. Introduction. CMAJ 160:837–839 [PMC free article] [PubMed] [Google Scholar]
- Ferguson RG (1955) Studies in tuberculosis. University of Toronto Press, Toronto [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros P, Skamene E, Forget A (1981) Genetic control of natural resistance to Mycobacterium bovis (BCG) in mice. J Immunol 127:2417–2421 [PubMed] [Google Scholar]
- Halpern J, Whittemore AS (1999) Multipoint linkage analysis: a cautionary note. Hum Hered 49:194–196 [DOI] [PubMed] [Google Scholar]
- Hodge SE, Abreu PC, Greenberg DA (1997) Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60:217–227 [PMC free article] [PubMed] [Google Scholar]
- Hodge SE, Elston RC (1994) Lods, wrods and mods: the interpretation of lod scores calculated under different models. Genet Epidemiol 11:329–342 [DOI] [PubMed] [Google Scholar]
- Hodge SE, Spence MA, Crandall BF, Sparkes RS, Sparkes MC, Crist M, Tideman S (1980) Huntington disease: linkage analysis with age-of-onset corrections. Am J Med Genet 5:247–254 [DOI] [PubMed] [Google Scholar]
- Holden M, Dubin MR, Diamond PH (1971) Frequency of negative intermediate-strength tuberculin sensitivity in patients with active tuberculosis. N Engl J Med 285:1506–1509 [DOI] [PubMed] [Google Scholar]
- Houston S, Fanning A, Soskolne CL, Fraser N (1990) The effectiveness of bacillus Calmette-Guerin (BCG) vaccination against tuberculosis: a case-control study in Treaty Indians, Alberta, Canada. Am J Epidemiol 131:340–348 [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Clark CD, Gschwend M, Justice-Higgins E (1997) PCR methods of genotyping. In: Dracopoli NC, Haines JL, Korf BR, Moir DT, Morton CC, Seidman CE, Seidman JG, et al (eds) Current protocols in human genetics. John Wiley & Sons, New York, pp 2.5.1–2.5.23 [DOI] [PubMed] [Google Scholar]
- Kallmann FJ, Reisner D (1943) Twin studies on the significance of genetic factors in tuberculosis. Am Rev Tuberculosis 47:549–574 [Google Scholar]
- Kruglyak L (1997) Nonparametric linkage tests are model free. Am J Hum Genet 61:254–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly ML, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, White RL (1986) Construction of human genetic linkage maps: likelihood calculations for multilocus analysis. Genet Epidemiol 3:39–52 [DOI] [PubMed] [Google Scholar]
- Lewis L-A, Victor TC, Hoal-van Helden EG, Blackwell JM, da Silva-Tatley F, Tullett S, Ehlers M, et al (1996) Identification of C to T mutation at position –236 bp in the human NRAMP1 gene promoter. Immunogenetics 44:309–311 [DOI] [PubMed] [Google Scholar]
- Liébana E, Aranaz A, Francis B, Cousins D (1996) Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J Clin Microbiol 34:933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J (1996) Molecular analysis of the telomeric half of human chromosome 2q. PhD thesis, Department of Experimental Medicine, McGill University, Montreal [Google Scholar]
- Liu J, Fujiwara TM, Buu NT, Sánchez FO, Cellier M, Paradis AJ, Frappier D, et al (1995a) Identification of polymorphisms and sequence variants in the human homologue of the mouse natural resistance-associated macrophage protein gene. Am J Hum Genet 56:845–853 [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stanton VP Jr, Fujiwara TM, Wang JX, Rezonzew R, Crumley MJ, Morgan K, et al (1995b) Large-scale cloning of human chromosome 2-specific yeast artificial chromosomes (YACs) using an interspersed repetitive sequences (IRS)-PCR approach. Genomics 26:178–191 [DOI] [PubMed] [Google Scholar]
- Mah MW, Fanning EA (1991) An epidemic of primary tuberculosis in a Canadian aboriginal community. Can J Infect Dis 2:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire W, Hill AVS, Allsopp CES, Greenwood BM, Kwiatkowski D (1994) Variation in the TNF-α promoter region associated with susceptibility to cerebral malaria. Nature 371:508–510 [DOI] [PubMed] [Google Scholar]
- Medina E, North RJ (1996) Evidence inconsistent with a role for the Bcg gene (Nramp1) in resistance of mice to infection with virulent Mycobacterium tuberculosis. J Exp Med 183:1045–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CG, May J, Stark K (1998) Human leukocyte antigens (HLA) in tuberculosis and leprosy. Trends Microbiol 6:148–154 [DOI] [PubMed] [Google Scholar]
- Miller MA (1991) A tuberculosis outbreak in a Native community: HLA linkage analysis and evaluation of diagnostic tests. MSc thesis, Department of Epidemiology and Biostatistics, McGill University, Montreal [Google Scholar]
- Morton NE (1998) Significance levels in complex inheritance. Am J Hum Genet 62: 690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North RJ, LaCourse R, Ryan L, Gros P (1999) Consequence of Nramp1 deletion to Mycobacterium tuberculosis infection in mice. Infect Immun 67:5811–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1991) Analysis of human genetic linkage, rev ed. Johns Hopkins University Press, Baltimore [Google Scholar]
- Risch N, Giuffra L (1992) Model misspecification and multipoint linkage analysis. Hum Hered 42:77–92 [DOI] [PubMed] [Google Scholar]
- Roth M-P, Nogueira L, Coppin H, Clanet M, Clayton J, Cambon-Thomsen A (1994) Tumor necrosis factor polymorphisms in multiple sclerosis: no additional association independent of HLA. J Neuroimmunol 51:93–99 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Schurr E, Skamene E (1995) The role of the Bcg gene in mycobacterial infection. In: Rom WN, Garay S (eds) Tuberculosis. Little, Brown, Boston, pp 247–259 [Google Scholar]
- Searle S, Blackwell JM (1999) Evidence for a functional repeat polymorphism in the promoter of the human NRAMP1 gene that correlates with autoimmune versus infectious disease susceptibility. J Med Genet 36:295–299 [PMC free article] [PubMed] [Google Scholar]
- Shaw MA, Collins A, Peacock CS, Miller EN, Black GF, Sibthorpe D, Lins-Lainson Z, et al (1997) Evidence that genetic susceptibility to Mycobacterium tuberculosis in a Brazilian population is under oligogenic control: linkage study of the candidate genes NRAMP1 and TNFA. Tuber Lung Dis 78:35–45 [DOI] [PubMed] [Google Scholar]
- Skamene E, Schurr E, Gros P (1998) Infection genomics: Nramp1 as a major determinant of natural resistance to intracelluar infections. Annu Rev Med 49:275–287 [DOI] [PubMed] [Google Scholar]
- Smith MHD, Marquis JR (1987) Tuberculosis and other mycobacterial infections. In: Feigin RD, Cherry JD (eds) Textbook of pediatric infectious diseases. WB Saunders, Philadelphia, pp 1342–1386 [Google Scholar]
- Stead WW (1997) The origin and erratic global spread of tuberculosis: how the past explains the present and is the key to the future. Clin Chest Med 18:65–77 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Ott J (1994) Handbook of human genetic linkage. Johns Hopkins University Press, Baltimore [Google Scholar]
- Udalova IA, Nedospasov SA, Webb GC, Chaplin DD, Turetskaya RL (1993) Highly informative typing of the human TNF locus using six adjacent polymorphic markers. Genomics 16:180–186 [DOI] [PubMed] [Google Scholar]
- Vidal SM, Malo D, Vogan K Skamene E, Gros P (1993) Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell 73:469–485 [DOI] [PubMed] [Google Scholar]
- Vidal S, Tremblay ML, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, et al (1995) The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med 182:655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AG, di Giovine FS, Blakemore AIF, Duff GW (1992) Single base polymorphism in the human tumour necrosis factor alpha (TNFa) gene detectable by NcoI restriction of PCR product. Hum Mol Genet 1:353 [DOI] [PubMed] [Google Scholar]