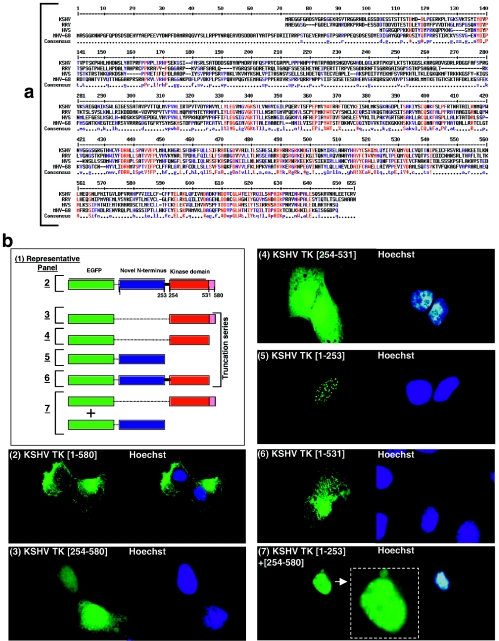
FIG.3.
Comparative alignment of the amino acid sequences of the N- and C-terminal domains of wild-type KSHV TK and related rhadinoviruses; generation and analysis of serial truncation mutants. (a) KSHV, RRV, HVS, and MHV-68 TKs were aligned using MultiAlign (9). (b) 143B TK- cells expressing EGFP-KSHV TK (WT) (panel 2) or truncation clones KSHV TK (254 to 580) (panel 3), KSHV TK (254 to 531) (panel 4), KSHV TK (1 to 253) (panel 5), KSHV TK (1 to 531) (panel 6), or a combination of both KSHV TK (1 to 253) plus KSHV TK (254 to 580) (panel 7). Cells counterstained with Hoechst are shown in the adjacent right-hand panels (blue staining).