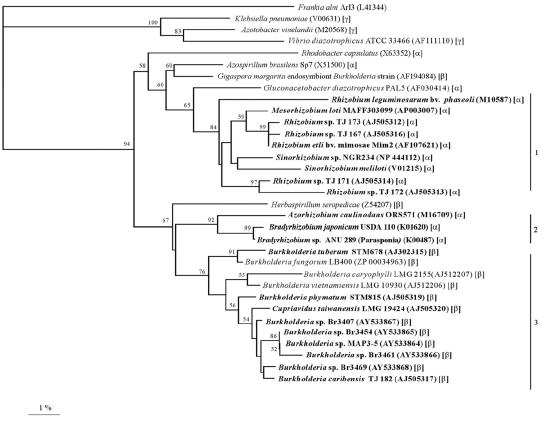
FIG. 5.
NifH phylogenetic tree. The tree was reconstructed by neighbor joining based on a 218-amino-acid alignment. Values along branches indicate bootstrap percentages of >50%, based on 1,000 replicates. The tree was rooted using sequences from Frankia alni, Vibrio diazotrophicus, Klebsiella pneumoniae, and Azotobacter vinelandii. Rhizobia are shown in bold, and the α-, β-, or γ-proteobacterial classification is indicated in parentheses. Clusters 1 and 2 contain α-rhizobia only, while cluster 3 includes both symbiotic and nonsymbiotic diazotrophic β-proteobacteria. nifH sequences for published bacteria are available from GenBank (accession numbers are given in parentheses).