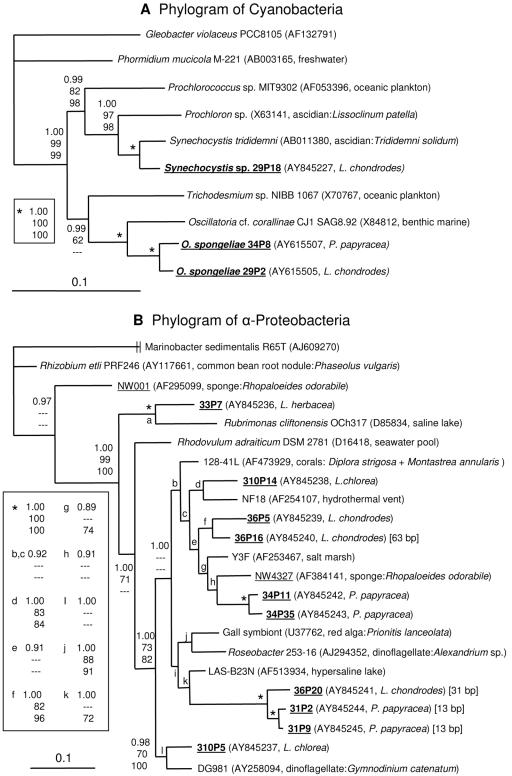
FIG. 4.
Phylogenetic trees generated with Mr. Bayes based on 16S rRNA gene sequences of cyanobacteria (A) and α-proteobacteria (B). Gleobacter violaceus is the outgroup in tree A, while the γ-proteobacterium Marinobacter sedimentalis is the outgroup in tree B. Sequences from sponges are underlined, and those from the Palauan sponges are in bold type. Next to the clone or culture designation in parentheses is given the GenBank accession number and the source of the sequence or culture. In the case of symbiotic bacteria, the host species is indicated. For the α-proteobacteria which contain insertions in the 16S rRNA gene, the size is indicated in brackets. Bootstrap values are shown only for branches which have a value over 60%, while Bayesian posterior probability is given for all branches (top, Bayesian posterior probability; middle, maximum parsimony bootstrap; bottom, neighbor joining bootstrap). A dash indicates a bootstrap value of less than 60% or clade not observed with that method. The scale bars indicate the number of substitutions per nucleotide position.