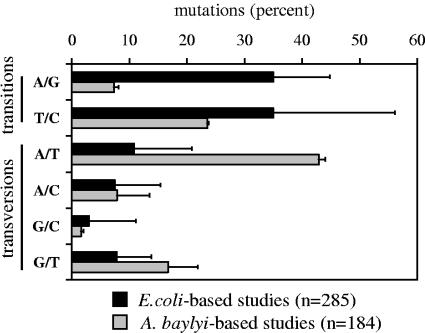
FIG. 1.
Distribution of single-nucleotide substitutions generated by a general random PCR mutagenesis approach (41). Natural transformation through which heteroduplex DNA was formed (25, 26) was used to obtain Acinetobacter mutants used in this analysis (15 [n = 76]; A. Buchan and L. N. Ornston, unpublished data [n = 108]). Mutants derived from E. coli-based approaches in which heteroduplex DNA is not formed were obtained from the following references: references 18 (n = 48), 30 (n = 14), 1 (n = 10), 36 (n = 35), 2 (n = 8), 33 (n = 9), 35 (n = 6), 42 (n = 8), 9 (n = 5), 19 (n = 18), 37 (n = 11), 40 (n = 13), 27 (n = 19), 4 (n = 5), and 20 (n = 76). If nucleotide substitutions are not explicitly described in the references, they were deduced from protein substitutions, and ambiguous substitutions were omitted. Error bars represent variations in frequencies among studies conducted with the two organisms. A. baylyi, Acinetobacter baylyi.