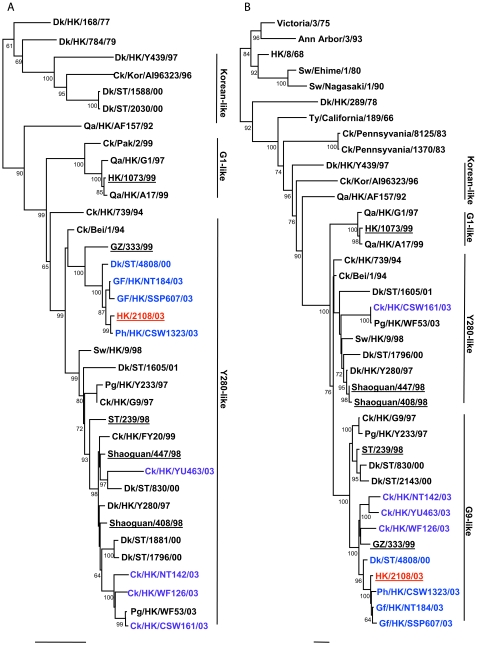
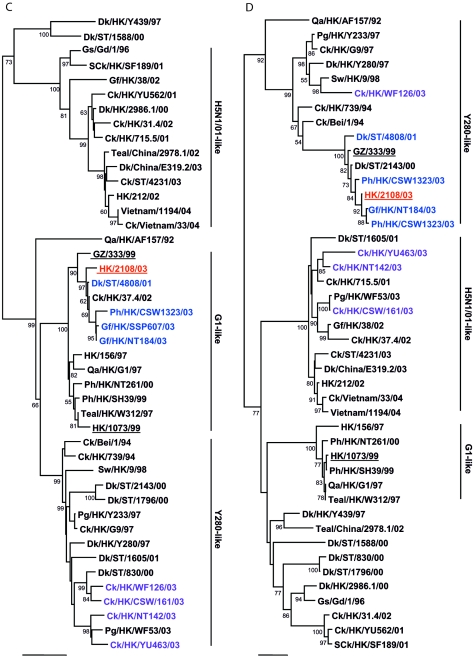
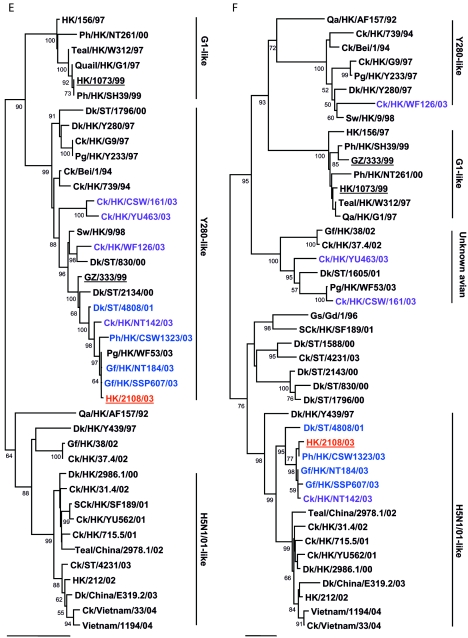
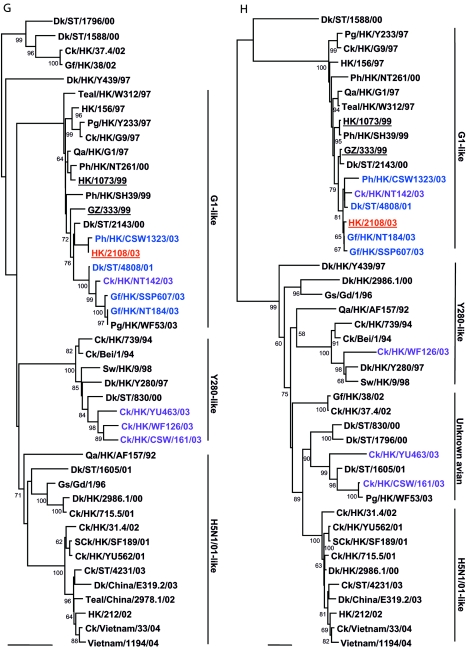
FIG.1.
Phylogenetic relationships of the influenza A virus genes HA, nucleotide positions 55 to 1042 (A); NA, nucleotide positions 1 to 1372 (B); M (matrix), nucleotide positions 1 to 972 (C); NP (nucleoprotein), nucleotide positions 49 to 1009 (D); NS (nonstructural), nucleotide positions 23 to 823 (E); PA (polymerase acidic), nucleotide positions 1423 to 2144 (F); PB1 (polymerase basic 1), nucleotide positions 1 to 1218 (G); and PB2 (polymerase basic 2), nucleotide positions 1101 to 2220 (H). Trees were generated with MEGA2 by using a Tamura-Nei (gamma) neighbor-joining analysis. The HA phylogenetic tree is rooted to A/turkey/Wisconsin/66; the NA tree is rooted to A/Leningrad/134/57; and the PA and PB1 trees are rooted to A/Ann Arbor/6/60 and B/Lee/40, respectively. The M, NP, NS, and PB2 trees are rooted to A/equine/Prague/1/56. Scale bars, 0.05 (HA, NS, PB1) and 0.02 (NA, M, NP, PA, PB2) nucleotide changes per nucleotide. The numbers at the nodes indicate bootstrap values from 500 bootstrap replicates. All viruses sequenced in the present study are underlined, and the remaining sequences can be found in GenBank. Abbreviations: Bei, Beijing; Cu, chukar; Ck, chicken; Dk, duck; GD, Guangdong; Gf, Guinea fowl; Gs, goose; GZ, Guangzhou; HK, Hong Kong; Kor, Korea; Pg, pigeon; Ph, pheasant; Qa, quail; SCk, silky chicken; ST, Shantou; Sw, swine; Ty, turkey; WDk, wild duck.