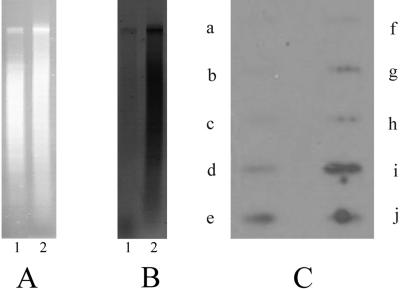
FIG. 3.
Southern and slot blot analyses using snRNA as a probe. (A) Ethidium bromide-stained agarose gel with total DNA (lane 1) and XbaI-digested micronuclear DNA (lane2). (B) Southern hybridization to total DNA (macronuclear and micronuclear DNA) from vegetative cells (lane 1) and to XbaI-digested micronuclear DNA (lane 2) using snRNAs from exconjugant cells at 10 h postconjugation as a probe. snRNA was isolated from the gel and end labeled as described previously (20). Hybridization was performed as described in Materials and Methods. (C) Slot blot of radiolabeled snRNAs of 10-h-postconjugation cells hybridized on different DNA sequences from Stylonychia: a, histone H4; b, calmodulin; c, rRNA genes; d, total DNA of vegetative cells; e, DNA isolated from polytene chromosomes; f, 1.3-kbp nanochromosome isolated from vegetative cells; g, pLJ01; h, 1.1-kbp and 1.3-kbp nanochromosome contained in pCE7; i, XbaI fragment of the repetitive element; j, MaA81. Each slot contains 1 μg DNA.