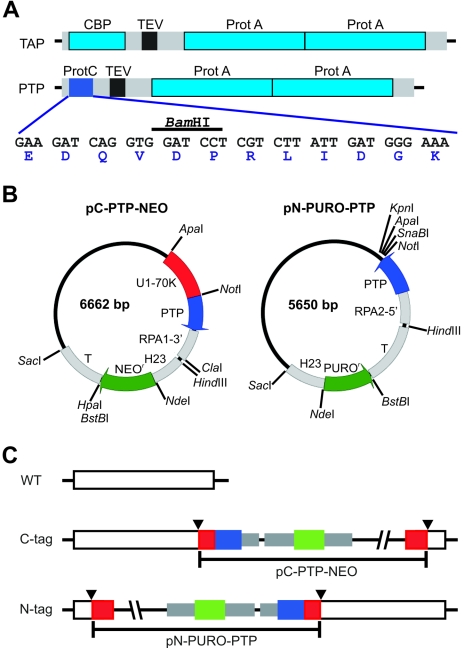
FIG. 1.
PTP tagging. (A) Schematic delineation (to scale) of the TAP and PTP tags. The ProtA and CBP epitopes of the original TAP tag are drawn in light blue, the TEV protease site is black, and spacer sequences are gray. In the PTP tag, the ProtC epitope is depicted in blue; its coding sequence, including a diagnostic BamHI restriction site and the amino acid sequence of the ProtC peptide, is provided below. (B) Circular map (to scale) of the T. brucei genome integration vectors pC-PTP-NEO and pN-PURO-PTP. Constructs are derivatives of pBluescript SK(+) and designed for genome integration. Each of them contains a PTP cassette for fusion of the PTP tag sequence to the target gene and a resistance marker cassette. The PTP sequence, T. brucei gene flanks, and the resistance marker (NEOr and PUROr) coding sequences are drawn in blue, gray, and green, respectively. Arrows indicate the direction of transcription. Most modules of the vectors can be individually excised by unique restriction sites as indicated. Gene flanks providing RNA processing signals for PTP fusion and resistance marker are the 3′ flank of TbRPA1 (RPA1-3′), HSP70 genes 2 and 3 intergenic region (H23), the β-α tubulin intergenic region (T), and the 5′ flank of TbRPA2 (RPA2-5′). While pC-PTP-NEO contains a target sequence (U1-70K, in red) which needs to be replaced, in pN-PURO-PTP a target sequence has to be inserted between the NotI and the following three restriction sites. (C) Illustration of PTP fusion to target genes. For PTP tagging of T. brucei proteins, pC-PTP-NEO and pN-PURO-PTP derivatives must be linearized inside the target sequence, depicted in red. Therefore, a prerequisite for targeted insertion of the constructs by homologous recombination is a unique restriction site within the target sequence (arrowheads). Colors are as above.