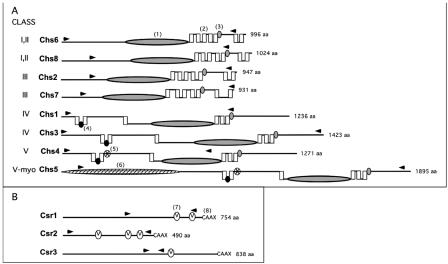
FIG. 2.
C. neoformans chitin synthase (Chs) and chitin synthase regulator (Csr) predicted protein structure comparison. The name of each protein is indicated to the immediate left of the protein structure. The predicted amino acid size (from the JEC21 genome sequence) is indicated to the right of each protein structure. A. Chitin synthases. Class designations are indicated by Roman numerals. Chs6 and Chs8 have protein characteristics of both class I and II. Chs5 (class V-myo) has class V protein sequence characteristics as well as a myosin motor domain. Domains were identified by rpsblast searches of the Conserved Domain Database at NCBI (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Domains are identified as follows: (1) large shaded ellipse, catalytic domain contains homologous sequence of chitin synthase domains 1 and 2 (pfam01644 and pfam03142); (2) vertical box, trans-membrane span; (3) small, shaded oval, conserved sequence “S/T-W-G-X-T/K-R/G”; (4) small black oval, conserved sequence “AWREK”; (5) small starred oval, cytochrome b5-like heme/steroid binding domain (pfam00173); (6) large hatched ellipse, myosin motor domain (cd 00124). B. Chitin synthase regulators. Domains are identified as (7) small “V” oval, SEL1 domain (smart 00671), which may be sites for protein-protein interaction; (8) C-terminal sequence “CAAX” as potential site for farensylation. The area between the arrow heads for each protein in both A and B indicates the region deleted in each deletion strain.