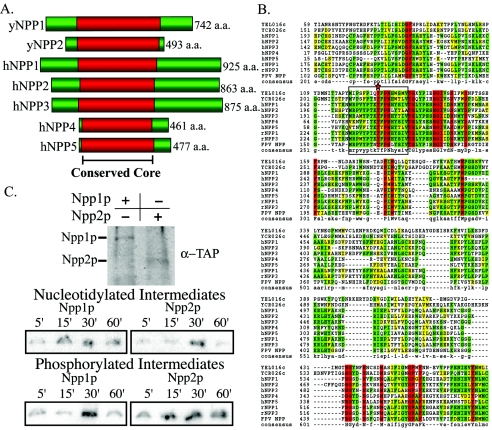
FIG. 2.
E-NPPs are conserved across species, including S. cerevisiae. (A) Alignment of two yeast genes with predicted NPPase activity. NPP1 and NPP2 were compared to members of the human E-NPP gene family. Alignments are centered on the conserved core residues (red) demonstrated to participate in catalytic activity in vertebrates. (B) Sequences of the two predicted E-NPPs in yeast are aligned with the five E-NPPs in H. sapiens (h, human), along with R. norvegicus (r, rat) and fowlpox virus (FPV, fowlpox virus) E-NPP genes. Red residues represent those conserved among all genes, green represents identical residues, and yellow represents similar residues between compared sequences. Boxed residues highlight the highly conserved active site of the E-NPP family, with the catalytic threonine residue is marked by a star. Protein sequences were retrieved from GenBank and were aligned with CLUSTALW. (C) Western analysis demonstrates isolation and purification of immunoprecipitated Npp1p and Npp2p TAP fusions utilized for the trapped intermediates and autophosphorylation experiments. Covalently trapped nucleotidylated intermediates were monitored over 60 min. Npp1p and Npp2p could perform the classic NPPase reaction where AMP was covalently attached to the catalytic threonine during catalysis. Autophosphorylation states were also monitored over 60 min as an indicator of autoinhibition. Npp2p was autophosphorylated at the earliest time point and persisted throughout the time course.