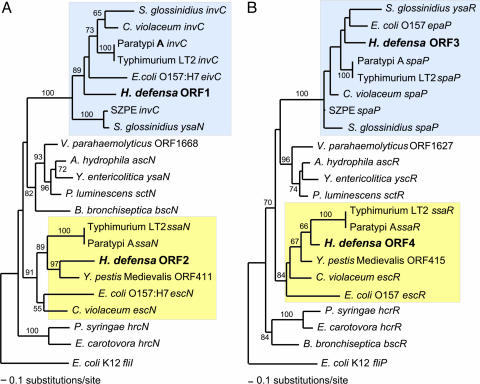
Fig. 1.
Phylogenetic analyses of homologs of genes encoding components of type III secretion systems (TTSS), including those newly discovered in H. defensa. (A) invC/ssaN. (B) spaP/ssaR; H. defensa possesses genes for two TTSS, corresponding to those specified within the SPI-1 and SPI-2 pathogenicity islands of Salmonella enterica (highlighted in blue and yellow, respectively). Identical results were obtained for homologs to spaQ/ssaS and to spaR/ssaT. Analyses were based on nucleotide alignments fit to alignments of inferred amino acid sequences by using CLUSTALW (63), followed by identification of initial maximum likelihood (ML) model of evolution (using AIC as implemented in MODELTEST version 3.16; ref. 64). ML heuristic searches were performed by using PAUP* version 4b10 (65) under the TBR branch-swapping option. Bootstrap support was estimated in parallel by using 100 searches and starting from 10 random trees.