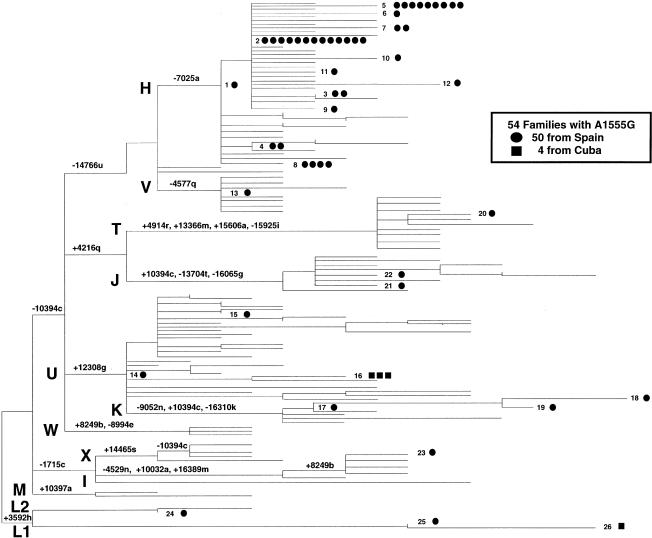
Figure 1.
Phylogenetic tree of RFLP haplotypes from patients with the deafness mutation A1555G. This mid-rooting-point maximum-parsimony tree includes 24 RFLP haplotypes (1–15 and 17–25) observed in 50 apparently unrelated Spanish patients (blackened circles), 2 RFLP haplotypes (16 and 26) observed in 4 Cuban patients (blackened squares), and the haplotypes previously described in 237 unrelated European controls. The capital letters H–M and T–X indicate haplogroups, and the numbers associated with the lowercase letters indicate the sites for restriction enzymes that define the specific haplogroups. The restriction enzymes are designated by single-letter code as follows: a = AluI; b = AvaII; c = DdeI; e = HaeIII; f = HhaI; g = HinfI; h = HpaI; k = RsaI; j = MboI; i = MspI; l = TaqI; m = BamHI; n = HaeII; q = NlaIII; r = BfaI; s = AccI; t = BstoI; u = MseI. The horizontal branch lengths are proportional to the number of mutational events that separate the haplotypes, with the exception of sites 10394c, 16303k, 16310k, and 16517e. In the parsimony analysis, these sites were assigned half the weight (i.e., 1) assigned to all other sites (i.e., 2). This tree is 430 steps in length and has consistency and retention indices of .652 and .896, respectively.