Summary
The clinical features of the 9p-deletion syndrome include dysmorphic facial features (trigonocephaly, midface hypoplasia, upward-slanting palpebral fissures, and a long philtrum) and mental retardation. The majority of these patients appear to have similar cytogenetic breakpoints in 9p22, but some cases show phenotypic heterogeneity. To define the breakpoints of the deleted chromosomes, we studied 24 patients with a deletion of 9p, by high-resolution cytogenetics, FISH with 19 YACs, and PCR using 25 different sequence-tagged sites. Of 10 different breakpoints identified, 9 were localized within an ∼5-Mb region, in 9p22-p23, that encompasses the interval between D9S1869 (telomeric) and D9S162 (centromeric). Eight unrelated patients had a breakpoint (group 1) in the same interval, between D9S274 (948h1) and D9S285 (767f2), suggesting a chromosome-breakage hotspot. Among 12 patients, seven different breakpoints (groups 3–9) were localized to a 2-Mb genomic region between D9S1709 and D9S162, which identified a breakpoint-cluster region. The critical region for the 9p-deletion syndrome maps to a 4–6-Mb region in 9p22-p23. The results from this study have provided insight into both the heterogeneous nature of the breakage in this deletion syndrome and the resultant phenotype-karyotype correlations.
Introduction
The chromosome-9p deletion was initially characterized in 1973 by Alfi et al., who reported a characteristic phenotype observed in six patients (Alfi et al. 1973) (monosomy 9p− syndrome [MIM 158170]). The major clinical features in this syndrome include mental retardation, trigonocephaly, midface hypoplasia, upward-slanting palpebral fissures, and a long philtrum (Young et al. 1982; Huret et al. 1988). Trigonocephaly and upward-slanting palpebral fissures are found in virtually all patients with this syndrome. In >100 patients, features frequently seen in this condition include hypertelorism, epicanthus, small palpebral fissures, flat nasal bridge, anteverted nares, low-set malformed posteriorly angulated ears, microstomia, micrognathia, short-appearing neck, widely spaced nipples, square hyperconvex nails, dolichomesophalangy, and hypotonia. Less frequently, other malformations have been seen, which include cardiac defects, hernias, omphaloceles, choanal atresia, abnormal genitalia, and scoliosis (Breg et al. 1976; Huret et al. 1988; Taylor et al. 1991; Bennett et al. 1993; Teebi et al. 1993; Shashi et al. 1994). Nonketotic hyperglycemia has been recognized in a small number of patients (Burton et al. 1989).
In a review of 80 literature cases with deletions of 9p, Huret et al. (1988) found that approximately half were due to de novo deletions and that the remaining half were due to unbalanced rearrangements. The majority (39/41) of the unbalanced rearrangements were familial. Cytogenetics studies have suggested that breaks can occur in 9p21-p23. To date, two studies have used additional molecular techniques to characterize a small number of cases (Teebi et al. 1993; Wagstaff and Hemann 1995). Teebi et al. (1993) characterized two rearranged chromosomes 9 by using chromosome-specific libraries. Wagstaff and Hemann (1995), utilizing microsatellite markers, characterized two deletions and initially reported the 9p deletion–syndrome critical region as including the interval between D9S286 and D9S162.
The lack of high-resolution chromosome analysis and molecular analysis can lead to both misassignment of the breakpoint and failure to detect additional chromosomal material or a derived chromosome 9 and can obscure phenotype/karyotype correlations. To define the breakpoints in all of the rearrangements in a large group of patients, we first determined whether these are deletions or cryptic unbalanced rearrangements and, second, established molecular breakpoints to determine the pattern and mechanism of breakage in these patients.
Patients and Methods
Clinical Population
A total of 24 patients with deletions or rearrangements in the short arm of chromosome 9 were ascertained for this study (table 1). Four of these samples (GM10994, GM01667, GM02356, and GM00870) were obtained from the National Institute of General Medical Sciences' Genetic Mutant Cell Repository. The other 20 samples were obtained directly from patients either seen at Case Western Reserve University and involved with the Chromosome 9p− Network or referred from other laboratories. All of these patients were reported to have trigonocephaly and features compatible with the 9p-deletion syndrome. The patient records for these latter 20 patients were reviewed by one of the authors (C.A.C.), to identify those clinical features of the 9p-deletion syndrome. Photographs were reviewed when patients were not available for physical examination.
Table 1.
Patient Population, Cytogenetic Abnormality, and Origin of Rearrangement
| Patient | Rearrangement (Type) | ParentalOrigin |
| Group 1: | ||
| 1a | Deletion (de novo) | Paternal |
| 2a | Deletion (de novo) | Paternal |
| 3 | Unbalanced translocationb (familial) | Maternal |
| 4a | Deletion (de novo) | Paternal |
| 5 | Deletion (de novo) | Unknown |
| 6 | Unbalanced translocationc (de novo) | Maternal |
| 7d | Deletion (unknown) | Unknown |
| 8 | Deletion (de novo) | Maternal |
| Group 1/2: | ||
| 9a | Deletion (de novo) | Paternal |
| Group 2: | ||
| 10a | Deletion (de novo) | Paternal |
| Group 3: | ||
| 11e | Deletion (unknown) | Unknown |
| 12a | Deletion (de novo) | Maternal |
| Group 4: | ||
| 13 | Unbalanced translocationf (familial) | Maternal |
| Group 5: | ||
| 14 | Deletion (de novo) | Paternal |
| Group 6: | ||
| 15a | Deletion (de novo) | Paternal |
| 16a | Inversion/deletiong (de novo) | Paternal |
| 17a | Deletion (de novo) | Maternal |
| Group 7: | ||
| 18h | Deletion (unknown) | Unknown |
| Group 8: | ||
| 19a | Deletion (de novo) | Maternal |
| 20a | Deletion (de novo) | Maternal |
| Group 8/9: | ||
| 21 | Duplication/deletioni (de novo) | Maternal |
| Group 9: | ||
| 22a | Deletion (de novo) | Maternal |
| Group 10: | ||
| 23a | Deletion (de novo) | Paternal |
| 24j | Deletion (de novo) | Unknown |
Reported by Micale et al. (1995).
46,XX,der(9)t(3;9)(p26.2;p22.3)mat.
46,XX,der(9)t(7;9)(p21.3;p22.1).
GM10094.
GM01667.
46,XY,der(9)t(3;9)(p24.2;p22.1)mat.
46,XX,del(9)(:q12→p22.3::q12→qter).
GM2356.
mos46,XX,del(9)(p22.1)/46,XX,der(9)t(9;9)(:p11→p22.1::p22.1→qter).
GM00870.
Cytogenetic Studies
Metaphase chromosomes were prepared either from phytohemagglutinin (PHA)-stimulated cultures or from lymphoblast or fibroblast cultures, by standard procedures. Leukocytes from peripheral blood of the probands were cultured to obtain chromosomes at ⩾600–750-band level (Yunis 1976; Ikeuchi 1984). PHA-stimulated blood leukocytes were cultured for ∼72 h in RPMI 1640 with 17% fetal bovine serum. The cultures were synchronized by the addition of thymidine for the last 16.5 h of culture, followed by the addition of ethidium bromide and colcemid for the last 45 min and 25 min of culture, respectively. The cells were treated for 8 min with 0.075 M KCl and were fixed in 3:1 methanol–acetic acid prior to staining. The chromosomes were GTG-banded, and ⩾20 chromosomal spreads were examined (Seabright 1971).
FISH Studies
FISH was performed with YACs and chromosome-specific paints on the unstained slides, according to the technique of Pinkel et al. (1986, 1988), with minor modifications (Sullivan et al. 1996). At least 10—and, in most cases, 20—metaphases were analyzed for the presence of probe on both the normal and the rearranged chromosome. All of the YACs utilized in this study were obtained from Research Genetics, after examination of the Whitehead Institute for Biomedical Research/MIT Center for Genome Research database. All of the YACs used in this study were mapped on the basis of sequence-tagged site (STS) content (fig. 1 and table 2).
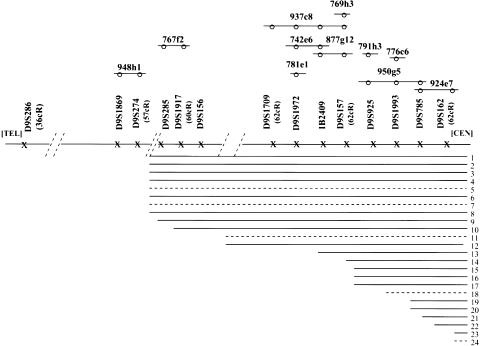
Figure 1.
YAC/STS map and 10 different breaks in 9p22-9p23 in the 24 patients studied. The STSs in our area of interest are ordered telomeric (TEL) to centromeric (CEN) on a horizontal axis, and several YACs localized to this region are placed above. The STSs contained in each YAC are indicated by circles. All of the STSs associated with the YACs used in the present study are listed; however, only a portion of these were used for breakpoint analysis and determination of parental origin (D9S268, D9S274, D9S285, D9S156, D9S157, D9S925, and D9S162). Only the YACs that were instrumental in the delineation of the groups (767f2, 948h1, 937c8, 742e6, 877g12, 769h3, 791h3, 950g5, and 924e7) are listed. Each patient is denoted by a line, indicating nondeleted material from chromosome 9p. Those patients studied by both YAC and microsatellite-marker analysis are denoted by a solid line, whereas patients studied only with YACs are denoted by a dashed line. The delineation of the various groups is shown in table 2. Patients 9 and 21 could not be precisely defined and were each placed within two groups (table 1).
Table 2.
Delineation of Breakpoint Groups
|
Status of YAC/STS |
||
| Group | Deleted | Present |
| 1 | 948h1 | 767f2 |
| 2 | D9S285 | V767f2 (reduced signal) |
| 3 | D9S156 | 742e6 |
| 4 | 718el | 769h3 |
| 5 | D9S157 | 877g2 |
| 6 | 769h3 | 791h3 |
| 7 | 791h3 | 776c5 |
| 8 | 776c5 | 950g5 |
| 9 | 950g5 | 924e7 |
| 10 | 924e7 | 778h8 |
Digital images were captured by a confocal microscope and/or Zeiss epifluorescent microscope equipped with a cooled CCD camera (Photometrics CG250) controlled by an Apple MacIntosh computer. Gray-scale source images were captured separately with DAPI, propidium iodide fluorescein, and rhodamine filter sets and were merged and pseudocolored by Gene Join software (Yale University).
Molecular Studies
Highly polymorphic microsatellite markers within or adjacent to the deleted region for each patient—and for their parents, when DNA from the latter was available (table 1)—were analyzed by PCR, by means of standard techniques. Primer sequences for microsatellite analysis (Operon Technologies or Research Genetics) were obtained from the Whitehead Institute for Biomedical Research/MIT Center for Genome Research and The Genome Database and were used with standard methods (Micale et al. 1995). Molecular distances in this study were estimated by radiation hybrid data from the Whitehead Institute for Biomedical Research/MIT Center for Genome Research.
Results
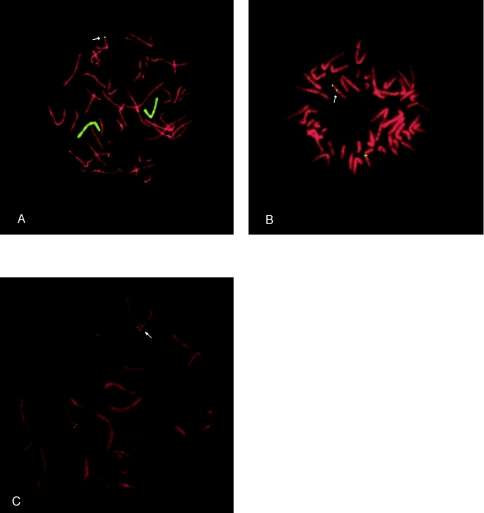
Initially, chromosome analyses of leukocytes at the 600–750-band level were performed; if the banding was suggestive of a rearrangement (e.g., in 3 of the 22 de novo cases), chromosome-specific paints were used to determine the origin of the extra material. Of the 24 cases examined in this study (table 1), 2 (cases 3 and 13) had a derived chromosome 9 due to malsegregation of a familial translocation. Of the other 22 cases studied, 19 appeared to be simple deletions; the remaining 3 cases involved more-complex rearrangements—a de novo duplication of 7p along with a 9p deletion, der(9)t(7;9)(p21.3;p22.1), in case 6; an inversion with a concomitant deletion, del(9)(:q12→p22.3::q12→qter), in case 16; and a mosaic intrachromosomal duplication and deletion of chromosome 9, mos46,XX,del(9)(p22.1)/46,XX,der(9)t(9;9)(:p11→p22.1::p22.1→qter), in case 21 (fig. 2). Chromosome analysis revealed that the breakpoints of the 24 patients were indistinguishable and within 9p22-p23. This finding suggested a need for more-detailed molecular cytogenetic studies involving both FISH with YACs and PCR analysis with microsatellite markers (figs. 1 and 3).
Figure 2.
FISH delineating the three structural abnormalities detected in this study. A, Chromosome 7 library, showing der(9)t(7;9) (case 6). B, Chromosome 9 classic satellite probe, showing the presence of an inversion in the inversion/deletion case (case 16). C, YAC 929d12, showing the duplication/deletion (case 21). Arrows indicate the derived chromosomes.
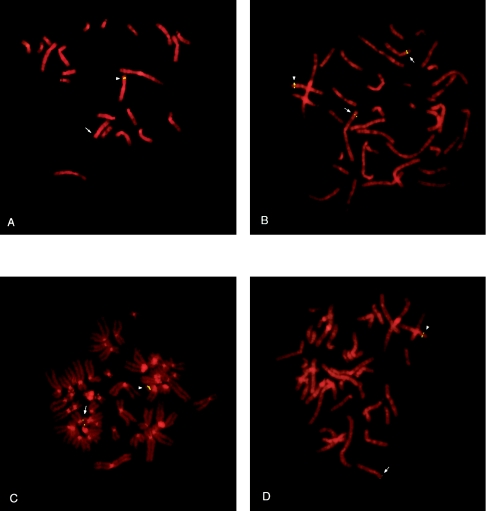
Figure 3.
Four examples of FISH with different YACs. A, Group 1 patient (case 8) with a deletion of YAC 948h1. The arrowhead indicates hybridization on the normal chromosome 9, and the arrow indicates lack of hybridization of the deleted chromosome 9. B, Group 6 translocation-carrier parent (case 3) with a split signal of YAC 937c8. The arrowhead indicates hybridization on the normal chromosome 9, and the arrow indicates hybridization on both the der(3) and the der(9). C, Group 2 patient (case 10) with a reduced signal from hybridization with YAC 767f2. The arrowhead indicates hybridization on the normal chromosome 9, and the arrow indicates a reduced signal on the deleted chromosome 9. D, Group 8 patient (case 20) with a reduced signal from hybridization YAC 950g5. The arrowhead indicates hybridization on the normal chromosome 9, and the arrow indicates a reduced signal on the deleted chromosome 9.
The results of our FISH analysis with YACs and microsatellite analysis indicate that various proximal breakpoints are involved in the deletions. At least 10 different molecular breakpoints (groups 1–10) were detected among the 24 patients (table 1 and fig. 1). Nine different breakpoints (groups 1–9) have been localized to an ∼5-Mb region in 9p22-p23, between D9S274 and D9S162 (fig. 1). All of these breakpoints were localized either within a YAC or between two STSs. Figure 1 illustrates the breakpoints identified in groups 1 and 2, between D9S274 and D9S285. PCR with microsatellite markers revealed that D9S274 was deleted in both groups, whereas D9S285 was present only in patient 10 (group 2), and FISH analysis demonstrated that sequences in YACs 948h1 and 924b12 were deleted in these patients, whereas those sequences were present in YAC 767f2. The patient in group 2 (patient 10) showed a reduced signal when hybridized with YAC 767f2 (fig. 3).
Eight of the 24 patients were determined to have the group 1 breakpoint, the largest number of patients assigned to any groups. Seven of the breakpoints (groups 3–9), encompassing 12 patients, were localized to just a 2-Mb region in 9p22 (fig. 1). All of these breaks were clustered between D9S1972 and D9S162, and the close proximity and number of breaks suggested a possible breakpoint-cluster region.
Parental Origin of the Deletion
Our initial studies delineating parental origin were performed in 13 families with de novo 9p deletions/rearrangements, by use of 9p dinucleotide-repeat microsatellite markers (Micale et al. 1995); we have now obtained an additional 4 families. Combined results from the 17 families have revealed that the 9p deletion is of paternal origin in 9 of them and is of maternal origin in 8 families.
Chromosome 9p Deletion: Critical Region
To better delineate a critical region for the 9p-deletion phenotype, we have correlated the above molecular findings with the phenotypic findings available for these patients. Records have been obtained on the available patients, although only a few have been studied clinically at Case Western Reserve University. Nonetheless, preliminary phenotype/karyotype correlation can be made on the basis of this information.
All of the patients with breaks in groups 1–10 show a characteristic phenotype compatible with the 9p-deletion syndrome. These features include trigonocephaly, flat occiput, small palpebral fissures, flat nasal bridge, anteverted nostrils, long philtrum, and microretrognathia. Recently, Wagstaff and Hemann's (1995) report of a patient with a 15-Mb interstitial deletion (from D9S286 to D9S162) helped to initially delineate this critical region. On the basis of our studies, we have reduced the size of the apparent critical region and can assign it to an ∼4–6-Mb region in 9p23—a region encompassing the interval between D9S286 and D9S285, the smallest region of overlap between our patients and the patient reported by Wagstaff and Hemann (1995).
Discussion
The chromosome 9–deletion syndrome is a fairly well-established and -recognized deletion syndrome (Huret et al. 1988). In this study we have examined 24 deletions or rearrangements of 9p, to delineate more precisely the nature of the abnormal chromosome and to determine the molecular breakpoints, using both FISH with single-copy probes and PCR with microsatellite markers.
Initial Cytogenetic Analysis
The majority of the cases obtained for this study had been diagnosed and analyzed previously. For consistency, each case, when possible, was restudied by high-resolution banding. Any detection of a banding pattern not consistent with a deletion was followed by use of a chromosome 9 library and subsequent use of other chromosome libraries, to detect the chromosomal origin of any non–chromosome 9 material. Three of the 22 de novo abnormalities were subjected to additional studies. These chromosomal and FISH studies not only detected 2 familial translocations and 19 de novo deletions but, in addition, also delineated 3 unusual de novo rearrangements (1 translocation, 1 inversion and deletion, and 1 intrachromosomal duplication and deletion). This encompassed ∼14% of the de novo rearrangements, a number higher than that shown in a previously published study (Huret et al. 1988). This is yet another example of how use of a combination of high-quality chromosome analysis and FISH can lead to the detection of an increased number of rearrangements and can improve the delineation of these abnormalities. Although there have been >100 cases of 9p deletions reported, previous cytogenetic studies have been limited in scope. Only 2 of the 80 cases (41 of which were de novo) reviewed by Huret et al. (1988) were due to de novo rearrangements. Our findings suggest that these rearrangements may occur at a frequency higher than previously reported and that the use of high-quality cytogenetic analysis with molecular cytogenetics is essential for the proper analysis of this group of abnormalities.
Breakpoint Analysis
Although >100 cases of 9p deletions have been reported (Huret et al. 1988; Burton et al. 1989; Taylor et al. 1991; Bennett et al. 1993; Teebi et al. 1993; Shashi et al. 1994; Wagstaff and Hemann 1995), the vast majority have been analyzed with only standard banding, yielding breakpoints in the range from 9p21 to 9p24. In our study, even with high-resolution chromosome analysis, it was difficult to differentiate the majority of the breakpoints. This is not surprising and is substantiated by numerous reports in the literature that have indicated the lack of a consensus breakpoint associated with the 9p-deletion syndrome.
Our study was undertaken to determine a consensus breakpoint; however, when a series of YACs and STSs were used, ⩾10 different breakpoints were identified (fig. 1). Eight (33%) of the breaks, the largest breakpoint group identified, were localized to a small region in 9p22 (group 1). Twelve other breaks (50%) were localized to a more proximal 2-Mb region, in 9p22, which included seven different breakpoints (groups 3–9; see fig. 1). These heterogeneous findings are surprising in view of the reports of site-specific breakpoints in several other deletion syndromes. A large number of deletion syndromes, especially contiguous-gene syndromes, have now been studied with detailed molecular techniques to elucidate breakpoints. Specific breakpoint sites have been found in >84% of the deletions seen in Smith-Magenis syndrome, and a novel junction fragment has been identified in 29 unrelated patients with Smith-Magenis syndrome (Juyal et al. 1996; Chen et al. 1997). Similar findings have been reported for Prader-Willi and Angelman syndromes, for DiGeorge syndrome, and for the 17p duplication and 17p deletion associated, respectively, with Charcot-Marie-Tooth (CMT) syndrome and hereditary neuropathy with liability to pressure palsies (HNPP) (Desmaze et al. 1993; Christian et al. 1995; Lupski et al. 1996). In all of these syndromes, deletions have been seen to occur at the same breakpoints, in the majority of the reported cases (Desmaze et al. 1993; Lindsay et al. 1995). Repetitive sequences have been implicated in the formation of the deletion in Prader-Willi and Angelman syndromes, and both repetitive sequences and the presence of a mariner transposon-like element have been implicated in the formation of both the deletion in HNPP and the duplication in CMT (Amos-Landgraf et al. 1994; Reiter et al. 1996; Kiyosawa and Chance 1996). In contrast to these studies, analysis of X isochromosomes and ring X chromosomes indicates not one site-specific break but, rather, a series of breaks within a small region (<5 Mb) (Wolff et al. 1994, 1996), similar to what has been seen in the present study. Additionally, studies of the formation of dicentric chromosome 15, inv dup(15), have shown several different areas of breakage (Leana-Cox et al. 1994).
It is not clear whether the deletions examined in the present study are interstitial or terminal. The cytological studies do not provide any obvious evidence that these are interstitial deletions; however, molecular methods must be used to detect and confirm cryptic rearrangements. Results of preliminary FISH experiments with YACs from 9p24 have also been compatible with a terminal versus an interstitial deletion (data not shown).
9p-Deletion Critical Region
In recent years, many different deletion syndromes have been examined to determine the minimal segment that is required to be deleted to produce a consensus phenotype. This has been most extensively studied in several contiguous-gene-deletion syndromes with microdeletions, such as Miller-Dieker syndrome, Smith-Magenis syndrome, Prader-Willi syndrome, and Angelman syndrome (Dobyns et al. 1993; Nicholls 1993; Buiting et al. 1995; Pilz et al. 1995; Juyal et al. 1996). In all of these syndromes, although studies are still ongoing, small regions have been implicated by molecular studies (Pilz et al. 1995; Juyal et al. 1996). This is also true for some of the larger-deletion syndromes, such as Wolf-Hirschhorn syndrome, cri-du-chat syndrome, Jacobsen syndrome, and 13q-deletion syndrome (Brown et al. 1993, 1995; Church et al. 1995; Estabrooks et al. 1995; Penny et al. 1995). Our studies have reduced the size of the critical region initially reported by Wagstaff and Hemann (1995) (i.e., from D9S286 to D9S162) and suggest that a critical region in the 9p-deletion syndrome can be assigned to an ∼4–6-Mb region from D9S286 to D9S285. All of the patients with a break centromeric to D9S274 (i.e., groups 1–10; see fig. 1) demonstrate the consensus 9p-deletion phenotype.
Parental Origin of the Deletion
Our studies have confirmed that the deletion/rearrangements in this syndrome can be of either maternal or paternal origin (Micale et al. 1995). This suggests that no known imprinted genes are involved in the pathogenesis of this syndrome, such as in Prader-Willi syndrome and Angelman syndrome (Nicholls 1993). Also, it agrees with previous studies indicating that no known imprinted genes are on human chromosome 9 (Ledbetter and Engel 1995).
Closer examination of these data does, however, reveal some intriguing points (table 1). When only deletions (not including either familial or de novo unbalanced rearrangements, since they may have indifferent underlying mechanisms) are viewed on the basis of the breakpoint group, the following results were seen: (1) four of five deletions in group 1 were paternal in origin; (2) three of the four deletions in groups 5 and 6 were paternal in origin; and (3) the deletions in group 8 were all maternal. Therefore, although paternal and maternal origin may be equally common overall, it is possible that the location of the breaks is correlated with parental origin.
In this study we have examined the origins and breakage in the 9p-deletion syndrome. Although these breaks have been found to occur in both maternal and paternal meiosis, no site-specific location of the breaks has been identified; rather, the breakage appears to be very heterogeneous, with 10 different breakpoint regions having been identified. Additional work will identify whether these breaks involve repetitive sequences of DNA, will address whether the breaks can be further subdivided as more genomic resources become available, and will allow more-complete phenotype-genotype correlations.
Acknowledgments
We wish to acknowledge the Chromosome 9p− Network and its assistance in patient recruitment for this study, and we thank J. Marie Haren for her technical assistance. This work was supported in part by a grant from the March of Dimes (Northern Ohio Division), by National Institutes of Health grant HD36252 (to S.S.), and by a grant from the Wilson Foundation to the Center for Human Genetics at University Hospitals of Cleveland and Rainbow Babies and Children's Hospital.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Chromosome 9p− Network, http://www.9pminus.org
- Genome Database, The, http://gdbwww.gdb.org (for primer sequences)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for monosomy 9p− syndrome [MIM 158170])
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www-genome.wi.mit.edu (for primer sequences, radiation-hybrid data, and YACs)
References
- Alfi O, Donnell GN, Crandall BF, Derencenyi A, Menon R (1973) Deletion of the short arm of chromosome 9 (46,9p−): a new deletion syndrome. Ann Genet 16:17–22 [PubMed]
- Amos-Landgraf J, Gottlieb W, Rogan PK, Nicholls RD (1994) Chromosome breakage in Prader-Willi and Angelman syndrome deletions may involve recombination between a repeat at the proximal and distal breakpoints. Am J Hum Genet Suppl 55:A38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CP, Docherty Z, Robb SA, Ramani P, Hawkins JR, Grant D (1993) Deletion 9p and sex reversal. J Med Genet 30:518–520 [DOI] [PMC free article] [PubMed]
- Breg WR, Aronson MM, Hill R, Green AE, Coriell LL (1976) Deletion in the short arm of chromosome 9 from a subject with congenital cerebral maldevelopment. Cytogenet Cell Genet 17:296–297 [DOI] [PubMed]
- Brown S, Gersen S, Anyane-Yeboa K, Warburton D (1993) Preliminary definition of a “critical region” of chromosome 13 in q32: report of 14 cases with 13q deletions and review of the literature. Am J Med Genet 45:52–59 [DOI] [PubMed]
- Brown S, Russo J, Chitayat D, Warburton D (1995) The 13q syndrome: the molecular definition of a critical deletion region in band 13q32. Am J Hum Genet 57:859–866 [PMC free article] [PubMed]
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B (1995) Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet 9:395–400 [DOI] [PubMed]
- Burton BK, Pettenati MJ, Block SM, Bensen J, Roach ES (1989) Nonketotic hyperglycemia in a patient with the 9p− syndrome. Am J Med Genet 32:504–505 [DOI] [PubMed]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, et al (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed]
- Christian SL, Robinson WP, Huang B, Mutirangura A, Line MR, Nakao M, Surti U, et al (1995) Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. Am J Hum Genet 57:40–48 [PMC free article] [PubMed]
- Church DM, Bengtsson U, Nielsen KV, Wasmuth JJ, Niebuhr E (1995) Molecular definition of deletions of different segments of distal 5p that result in distinct phenotypic features. Am J Hum Genet 56:1162–1172 [PMC free article] [PubMed]
- Desmaze C, Prieur M, Amblard F, Aïkem M, LeDeist F, Demczuk S, Zucman J, et al (1993) Physical mapping by FISH of the DiGeorge critical region (DGCR): involvement of the region in familial cases. Am J Hum Genet 53:1239–1249 [PMC free article] [PubMed]
- Dobyns WB, Reiner O, Carrozzo R, Ledbetter DH (1993) Lissencephaly: a human brain malformation associated with deletion of the LIS1 gene located at chromosome 17p13. JAMA 270:2838–2842 [DOI] [PubMed]
- Estabrooks LL, Breg WR, Hayden MR, Ledbetter DH, Myers RM, Wyandt HE, Yang-Feng TL, et al (1995) Summary of the 1993 ASHG ancillary meeting “Recent Research on Chromosome 4p Syndromes and Genes.” Am J Med Genet 55:453–458 [DOI] [PubMed]
- Huret JL, Leonard C, Forestier B, Rethore MO, Lejeune J (1988) Eleven new cases of del(9p) and features from 80 cases. J Med Genet 25:741–749 [DOI] [PMC free article] [PubMed]
- Ikeuchi T (1984) Inhibitory effect of ethidium bromide on mitotic chromosome condensation and its application to high resolution chromosome banding. Cytogenet Cell Genet 38:56–61 [DOI] [PubMed]
- Juyal RC, Figuera LE, Hauge X, Elsea SH, Lupski JR, Greenberg F, Baldini A, et al (1996) Molecular analyses of 17p11.2 deletions in 62 Smith-Magenis syndrome patients. Am J Hum Genet 58:998–1007 [PMC free article] [PubMed]
- Kiyosawa H, Chance PF (1996) Primate origin of the CMT1A-REP repeat and analysis of a putative transposon-associated recombinational hotspot. Hum Mol Genet 5:745–753 [DOI] [PubMed]
- Leana-Cox J, Jenkins L, Palmer CG, Plattner R, Sheppard L, Fletjer WL, Zackowski J, et al (1994) Molecular cytogenetic analysis of inv dup(15) chromosomes using probes specific for the Prader-Willi/Angelman syndrome region: clinical implications. Am J Hum Genet 54:748–756 [PMC free article] [PubMed]
- Ledbetter DH, Engel E (1995) Uniparental disomy in humans: development of an imprinting map and its implications for prenatal diagnosis. Hum Mol Genet 4:1757–1764 [DOI] [PubMed]
- Lindsay EA, Greenberg F, Shaffer LG, Shapira SK, Scambler PJ, Baldini A (1995) Submicroscopic deletions at 22q11.2: variability of the clinical picture and delineation of a commonly deleted region. Am J Med Genet 56:191–197 [DOI] [PubMed]
- Lupski JR, Roth JR, Weinstock GM (1996) Chromosomal duplications in bacteria, fruit flies, and humans. Am J Hum Genet 58:21–27 [PMC free article] [PubMed]
- Micale MA, Haren JM, Conroy JM, Crowe CA, Schwartz S (1995) Parental origin of de novo chromosome 9 deletions in del(9p) syndrome. Am J Med Genet 57:79–81 [DOI] [PubMed]
- Nicholls RD (1993) Genomic imprinting and uniparental disomy in Angelman and Prader-Willi syndrome: a review. Am J Med Genet 46:16–25 [DOI] [PubMed]
- Penny LA, Dell'Aquila M, Jones MC, Bergoffen J, Cunniff C, Fryns J-P, Grace E, et al (1995) Clinical and molecular characterization of patients with distal 11q deletions. Am J Hum Genet 56:676–683 [PMC free article] [PubMed]
- Pilz DT, Dalton A, Long A, Jaspan T, Maltby EL, Quarrell OWJ (1995) Detecting deletions in the critical region for lissencephaly on 17p13.3 using fluorescent in situ hybridization and a PCR assay identifying a dinucleotide repeat polymorphism. J Med Genet 32:275–278 [DOI] [PMC free article] [PubMed]
- Pinkel D, Landegent J, Collins C, Fusco J, Segraves R, Lucas J, Gray JW (1988) Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA 85:9138–9142 [DOI] [PMC free article] [PubMed]
- Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938 [DOI] [PMC free article] [PubMed]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 [DOI] [PubMed]
- Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2:971–972 [DOI] [PubMed]
- Shashi V, Golden WL, Fryburg JS (1994) Choanal atresia in a patient with the deletion (9p) syndrome. Am J Med Genet 49:88–90 [DOI] [PubMed]
- Sullivan BA, Jenkins LS, Karson EM, Leana-Cox J, Schwartz S (1996) Evidence for structural heterogeneity from molecular cytogenetic analysis of dicentric Robertsonian translocations. Am J Hum Genet 59:167–175 [PMC free article] [PubMed]
- Taylor LD, Krizman DB, Jankovic J, Hayani A, Steuber PC, Greenberg F, Fenwick RG, et al (1991) 9p Monosomy in a patient with Gilles de la Tourette's syndrome. Neurology 41:1513–1515 [DOI] [PubMed]
- Teebi AS, Gibson L, McGrath J, Meyn MS, Breg WR, Yang-Feng TL (1993) Molecular and cytogenetic characterization of 9p− abnormalities. Am J Med Genet 46:288–292 [DOI] [PubMed]
- Wagstaff J, Hemann M (1995) A familial “balanced” 3;9 translocation with cryptic 8q insertion leading to deletion and duplication of 9p23 loci in siblings. Am J Hum Genet 56:302–309 [PMC free article] [PubMed]
- Wolff DJ, Brown CJ, Schwartz S, Duncan AMV, Surti U, Willard HF (1994) Small marker X chromosomes lack the X inactivation center: Implications for karyotype/phenotype correlations. Am J Hum Genet 55:87–95 [PMC free article] [PubMed]
- Wolff DJ, Miller AP, Van Dyke DL, Schwartz S, Willard HF (1996) Molecular definition of breakpoints associated with human Xq isochromosomes: implication for mechanisms of formation. Am J Hum Genet 58:154–160 [PMC free article] [PubMed]
- Young RS, Reed T, Hodes ME, Palmer CG (1982) The dermatoglyphics and clinical features of the 9p trisomy and partial 9p monosomy syndromes. Hum Genet 62:31–39 [DOI] [PubMed]
- Yunis J (1976) High resolution of human chromosomes. Science 191:1268–1270 [DOI] [PubMed]