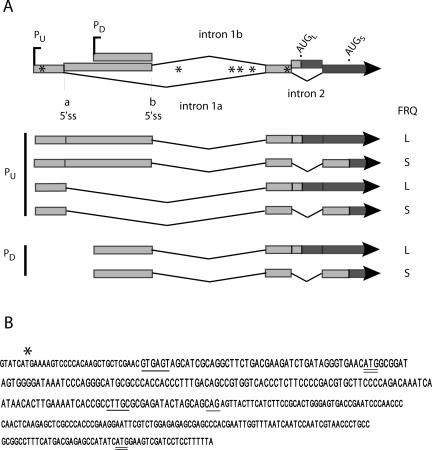
Figure 1.
Schematic of frq transcript structure. (A) This diagram is a compilation of the results from many experiments (see below), and represents the first ca. 1900 nt of the transcribed region of frq. The alternative start sites are designated PU and PD. The broad Vs represent introns. Alternative 5′ splice sites for the large upstream intronic region are called a and b. Intron 2 encompasses AUGL, as shown. The asterisks (*) represent upstream AUGs. The lighter shading indicates exon sequences, which vary among the transcripts, and the darker shading denotes coding sequences. (B) The sequence of intron 2 and adjacent regions is shown. Consensus sequences for splicing are underlined; the 5′ and 3′ splice sites were determined experimentally, but the branchpoint sequence shown, CTTGC, is the closest match to the known consensus for Neurospora (see Results). Initiation codons for LFRQ and SFRQ are doubly underlined, and the ATG for uORF 6 is indicated by an asterisk.