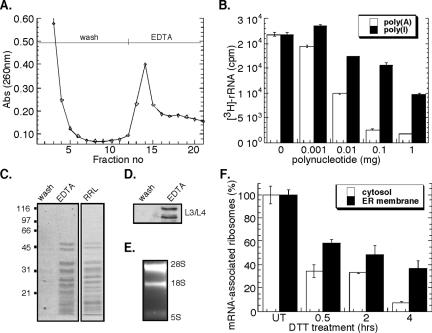
Figure 4.
Poly(U)-Sepharose affinity chromatographic analysis of mRNA-associated ribosomes. Native J558L ribosome-mRNA complexes were affinity purified by poly(U)-Sepharose resin. (A) Chromatogram of ribosome elution by EDTA was monitored by UV absorbance (260 nm). Competitive inhibition of metabolically labeled 3H-ribosome binding using the polynucleotides poly(A) or poly(I) (B). Coomassie blue staining of SDS-PAGE profile of EDTA-eluted fractions compared with rabbit reticulocyte-derived ribosomes (C). Immunoblot analysis of L3/L4 ribosomal proteins from wash versus EDTA-eluted fractions (D). Ethidium bromide-stained denaturing agarose gel of EDTA-eluted fractions (E). J558L cells were treated with 10 mM DTT for the indicated times. The mRNA occupancy levels of cytosolic and ER-derived, metabolically labeled 3H-ribosomal fractions were determined from the EDTA-eluted fractions. mRNA (100%) association corresponds to the occupancy level obtained in untreated cells (t = 0 h) (F).