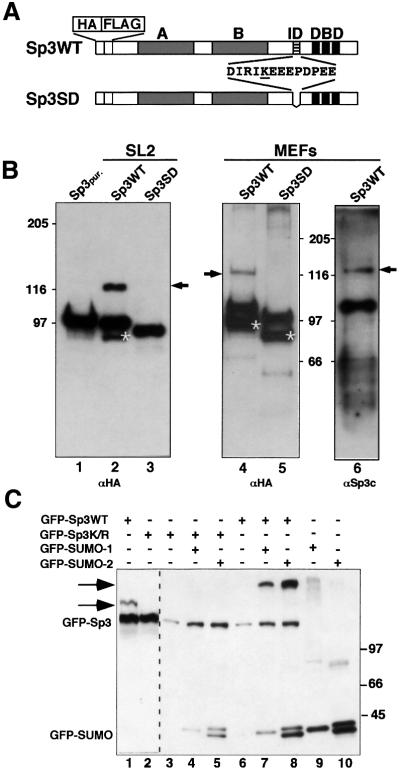
Fig. 1. Sp3 is SUMO modified in vivo. (A) Schematic representation of HA- and FLAG-tagged wild-type Sp3 protein Sp3WT and the Sp3 mutant Sp3SD expressed in SL2 cells (Braun and Suske, 1999) and in MEFs deficient of endogenous Sp3 (H.Göllner and G.Suske, unpublished data). Grey boxes indicate the two glutamine-rich activation domains A and B, and three black stripes the zinc fingers of the DNA-binding domain (DBD) of Sp3. The ID of Sp3 is depicted by hatched stripes. Amino acids that are deleted in the Sp3SD mutant protein are shown. The single lysine within this sequence is underlined. (B) Western blot analyses of epitope-tagged Sp3WT and mutant Sp3SD protein. HA- and FLAG-tagged Sp3WT- or Sp3SD-expressing SL2 cells (lanes 2 and 3) and MEFs (lanes 4, 5 and 6) were lysed with SDS-containing buffer. Proteins were separated on 7.5% SDS– polyacrylamide gels and blotted to PVDF membranes. Membranes were subsequently incubated with HA- (αHA) or Sp3-specific (αSp3c) antibodies as indicated. Arrows point to the covalently modified wild-type Sp3WT protein. Asterisks depict C-terminally deleted degradation products of Sp3 and Sp3SD that were detectable with the αHA antiserum but not with the αSp3c antibodies that recognize an epitope at the Sp3 C-terminal end. Lane 1 contains affinity-purified epitope-tagged Sp3 protein (Sp3pur.) (Braun et al., 2001) lacking the covalent modification. (C) GFP fusion vectors (3 µg) for GFP–Sp3WT, GFP–Sp3K/R, GFP–SUMO-1 and GFP–SUMO-2 were transiently transfected in Ishikawa cells as indicated. Cells were lysed and equal amounts of total cellular proteins (20 µg per lane) were separated on a 6.0% SDS–polyacrylamide gel and blotted to PVDF membranes. Detection was by immunoblotting with αGFP antibodies. The arrows point to SUMO-modified GFP–Sp3WT. The occurrence of two GFP–SUMO-2 forms is most likely due to incomplete processing.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.