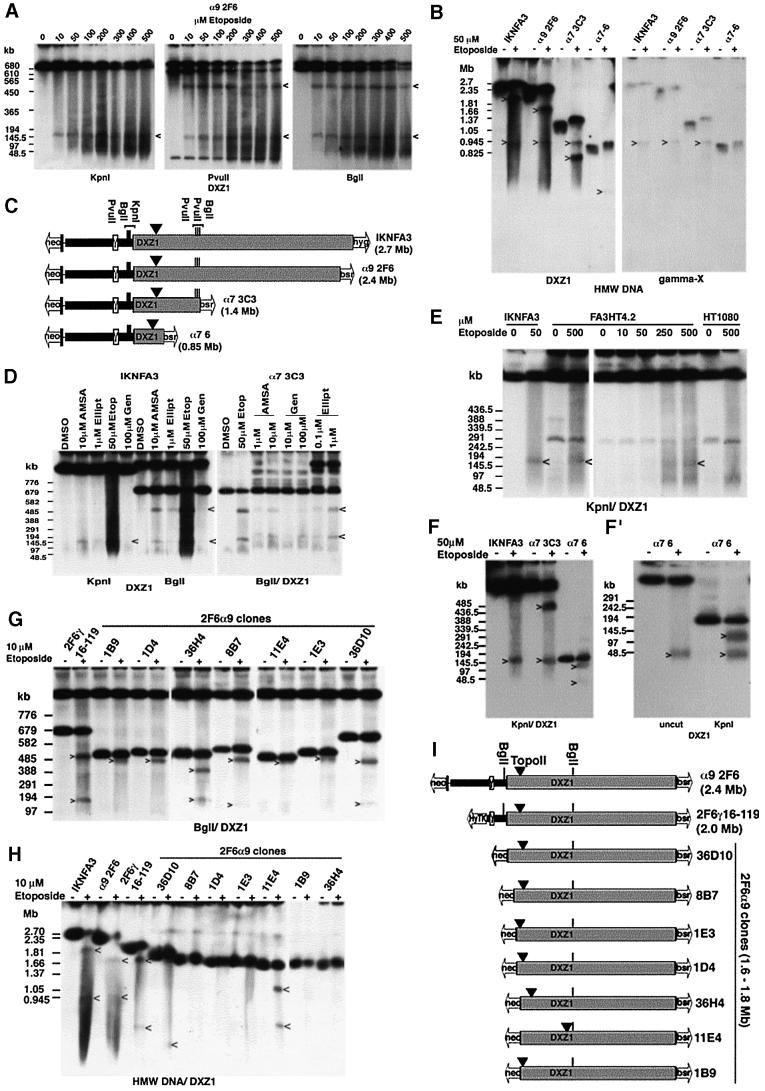
Fig. 7. Mapping topo II activity within the DXZ1 array. (A) DT40 cells carrying the α9 2F6 minichromosome were incubated with etoposide in vivo for 30 min and embedded in agarose. HMW DNA was cut with KpnI, PvuII or BglI, resolved by PFGE and Southern blotted with DXZ1. Etoposide-specific DXZ1-hybridizing fragments are indicated. PFGE conditions: 20–60 s pulse, 200 V, 24 h. (B) Uncut HMW DNA from various 50 µM etoposide-treated (+) or DMSO only (–) minichromosome-carrying DT40 lines from the Xq-truncation series was resolved by PFGE. The Southern blot filter was hybridized sequentially for X γ-satellite (gamma-X) and DXZ1. Etoposide-specific hybridizing fragments are indicated. (The reason for the slight mobility shift of the intact minichromosomes following etoposide exposure is unknown, but may be due to an effect of the apoptotic genome-wide chromosome fragmentation on HMW DNA migration conditions.) (C) Schematic showing localization of the topo II cleavage site on IKNFA3 and various minichromosomes from the Xq-truncation series. Also indicated are the positions of BglI, PvuII and KpnI sites within and flanking the DXZ1 array (grey box). (D) The IKNFA3 and α7 3C3 DT40 cell lines treated with various topo II inhibitors: AMSA (amsacrine), Ellipt (ellipticine), Etop (etoposide) and Gen (genistein), or with DMSO only. HMW DNA was digested with BglI or KpnI, resolved by PFGE and Southern blotted with DXZ1. PFGE conditions: 40–80 s pulse, 200 V, 24 h. The 520 and 150 kb inhibitor-specific cleavage products are indicated. (E) Preservation of the position of topo II cleavage on the IKNFA3 minichromosome in human and chicken. FA3HT4.2 (minichromosome in HT1080), HT1080 and IKNFA3 (minichromosome in DT40) were treated with etoposide for 60 min before preparation for PFGE. KpnI-cut HMW DNA was Southern blotted with DXZ1. PFGE conditions: 20–60 s pulse, 200 V, 24 h. Etoposide-specific DXZ1-hybridizing KpnI fragments are indicated. (F and F′) Analysis of etoposide-sensitive topo II cleavage in the α7 6 minichromosome using KpnI-cut and uncut HMW DNA. Samples are 50 µM etoposide-treated (+) or DMSO only (–). PFGE conditions for (F): 20–60 s pulse, 200 V, 24 h and for (F′): 10–30 s pulse, 200 V, 18 h in 1.5% agarose. Etoposide-specific DXZ1-hybridizing fragments are indicated. (G) Analysis of topo II cleavage in various 2F6α9 clones from the Xp- truncation series. HMW DNA from 10 µM etoposide-treated (+) and untreated (–) cells was digested with BglI, resolved by PFGE and Southern blotted. Etoposide-specific DXZ1-hybridizing fragments are indicated. PFGE conditions were 40–80 s pulse at 200 V for 24 h. (H) Analysis of topo II cleavage in various 2F6α9 clones from the Xp-truncation series. Uncut HMW DNA from etoposide-treated (+) and untreated (–) cells was resolved by PFGE and the Southern blotted for DXZ1. Etoposide-specific hybridizing fragments are indicated. (I) Schematic showing localization of topo II cleavage. The position of BglI sites relative to the DXZ1 array (grey box) is indicated.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.