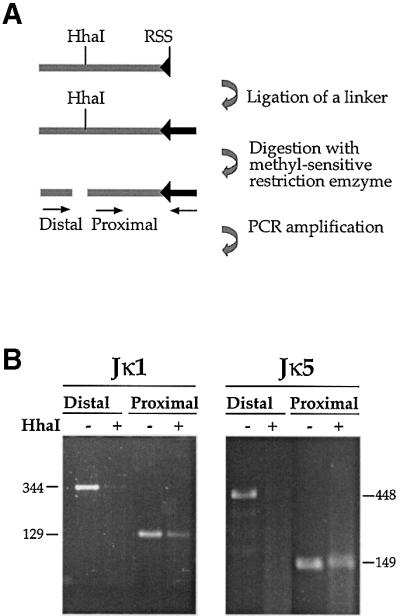
Fig. 3. Pre-existing DSBs in Jκ. (A) Strategy of methylation-sensitive LM-PCR analysis. CD19+ cells were isolated from BM of wild-type mice. These cells actively rearrange the κ locus and therefore contain signal end intermediates which can be detected by ligation of total genomic DNA to a partially double-strand oligonucleotide linker (the darker bar attached to RSS). Following ligation, half of the sample was digested with the methylation-sensitive restriction enzyme, HhaI, and both the digested and undigested samples were PCR analyzed with two sets of primers. In each set, one primer is specific for the linker and the other for the Jκ locus. The proximal set uses a primer located downstream of the HhaI site and thus yields a PCR product irrespective of the methylation state. The distal set contains a primer flanking the HhaI site, making this PCR reaction sensitive to methylation. (B) Genomic DNA purified from CD19+ BM cells of wild-type mice was linker- ligated and analyzed by LM-PCR for DSBs at RSSs flanking the Jκ1 and Jκ5 gene segments (see map in Figure 2).
