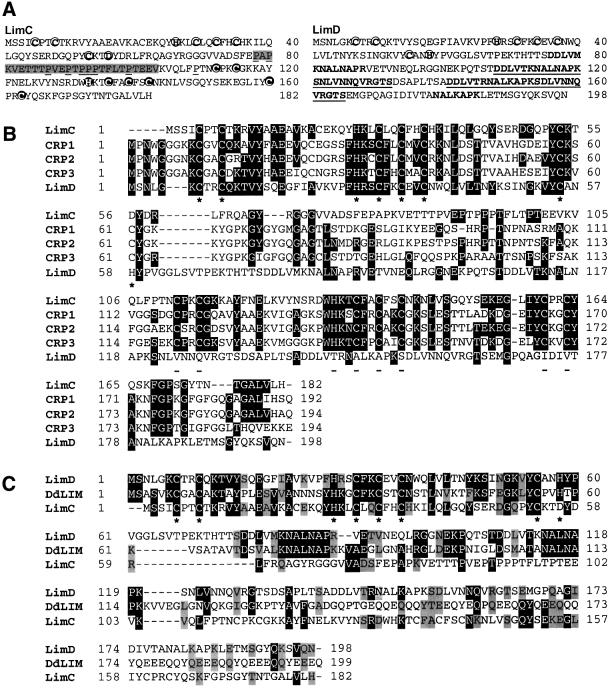
Fig. 1. (A) Deduced amino acid sequence of LimC and LimD. The conserved cysteine, histidine or aspartate residues of the N-terminal LIM domains of LimC and LimD (bold, circled letters) and the C-terminal LIM domain of LimC (bold, shaded and circled letters) are shown. The intervening sequence between the two LIM domains of LimC exhibiting low compositional complexity is shaded, the proline residues are underlined. The two nearly perfect repeat motifs of 25 amino acid residues (shown in bold, underlined letters) and two shorter versions of this repeat motif (shown in bold letters) in the C-terminal region of LimD are indicated. (B) Alignment of the LimC and LimD amino acid sequences with members of the CRP family, CRP1, CRP2 and CRP3. Residues that are identical between LimC and/or LimD and any of the other members of the CRP family are shadowed in dark gray. Asterisks and dashes below the aligned sequences indicate the N-terminal LIM consensus (in all sequences) and C-terminal LIM consensus (in all sequences except LimD), respectively. (C) Alignment of the LimC and LimD amino acid sequences with Dictyostelium DdLim. Residues that are identical between any two sequences are shadowed in dark gray, and residues that are similar are shadowed in gray. Asterisks below the aligned sequences indicate the N-terminal LIM consensus. Sequences were aligned using the Clustal_W program (Wisconsin package version 9.0). Dashes indicate gaps introduced in the sequence for optimal alignment. DDBJ/EMBL/GenBank accession Nos: LimC, AF348466; LimD, AF348467; DdLim, U97699.1; CRP1, P32965; CRP2, P50460; CRP3, P50463.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.