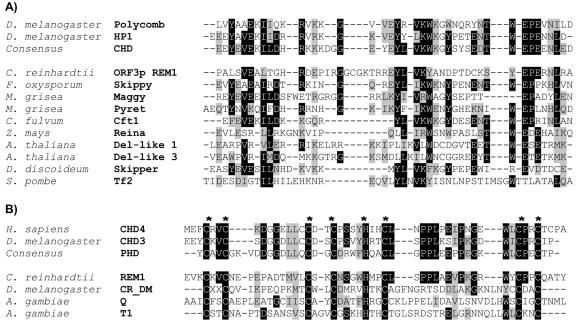
FIG. 4.
Alignment of chromodomains and PHD module from chromatin modifiers proteins and various gypsy-type LTR retrotransposons. (A) The upper group includes authentic chromodomains of chromatin-modifying proteins polycomb (P26017) and HP1 (P05205) of Drosophila melanogaster and the consensus sequence of chromodomain available at the NCBI conserved domain database. The lower group represents chromodomains of CrREM1 and 3′-end integrase of Ty3-gypsy class of LTR retrotransposons. The accession numbers of retrotransposons which have been translated from their DNA sequence are as follows: ORF3p REM1, AA073552; Reina, U69258 and Pyret, AB062507. The accession numbers of retrotransposons available in the protein databases are as follows: Del-like 1 and 3, AC002534 and Z97342; Cft1, AF051915; Maggy, AAA33420; Skippy, AAA88791; Tf2, T38401; and Skipper, AAC48336. (B) The upper block includes authentic PHD-finger domain from chromatin-modifying proteins: chromo domain helicase-DNA-binding protein 4 (CHD4) of Homo sapiens (Q14839), CHD3 of Drosophila melanogaster (AAF49162), and consensus sequence of PHD available in the NCBI conserved domain database. The lower block contains PHD conserved in ORF3p encoded by REM1 of C. reinhardtii (AA073552), ORF1p encoded by non-LTR retrotransposons (CR_DM of Drosophila melanogaster, [15]), and the retrotransposons Q (T43019) and T1 (M93689) of Anopheles gambiae. The asterisks show conserved cysteine and histidine residues matching the PHD domain. Black boxes, identical amino acids; gray boxes, similar amino acids.