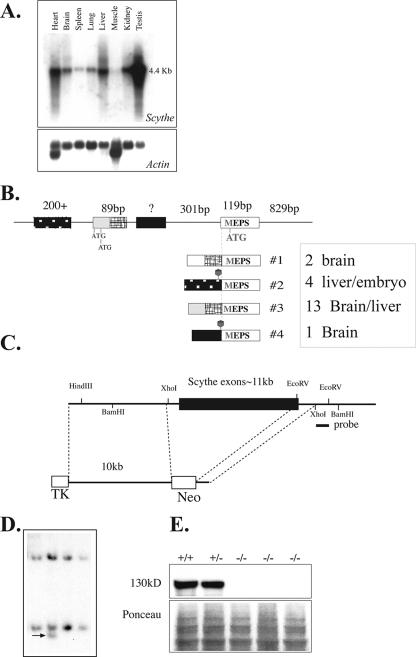
FIG. 1.
Inactivation of mouse Scythe. (A) Northern blot analysis of Scythe mRNA expression in various adult tissues (top). Actin reprobing of the blot confirmed equal RNA loading (bottom). (B) Schematic representation of the Scythe 5′ UTR sequence and the alternative splices found by reverse transcriptase PCR for several tissues, including liver and brain. The sizes of some 5′ UTR and coding exons are shown schematically positioned on genomic DNA, and the spliced transcripts are indicated below the genomic DNA structure. The relative abundance of a particular transcript is indicated by the number of times it was isolated by PCR from a particular tissue, i.e., transcript no. 3 was the most common 5′ version of the Scythe. (C) Structure of the Scythe targeting vector that replaced almost the entire Scythe ORF. The probe used for the Southern blot analysis is indicated. TK, thymidine kinase. (D) Southern blot analysis after BamHI digestion of ES cells. (E) Scythe (130 kDa) is present in WT (+/+) and heterozygous (+/−) cells but is absent in Scythe-null (−/−) mouse embryo fibroblasts.