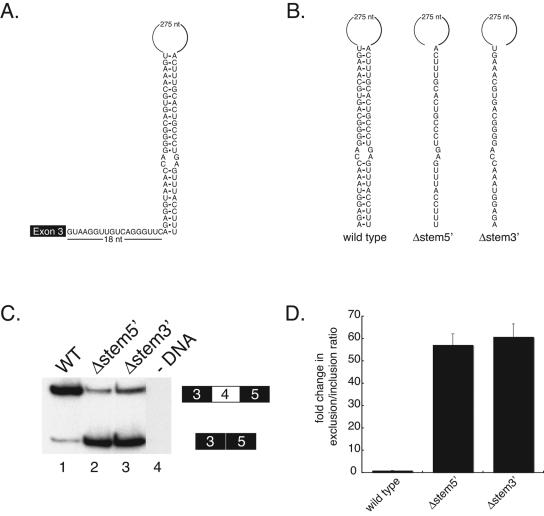
FIG. 2.
Disruption of a putative RNA secondary structure results in exon 4 skipping. (A) Diagram of the potential RNA secondary structure formed by base pairing of sequences 18 nt downstream of exon 3 with a sequence in the region identified in Fig. 1. (B) Diagram of the locations of deletions made within the stem of the putative RNA secondary structure element. WT, wild type. (C) Polyacrylamide gel of RT-PCR products from RNA isolated from Drosophila S2 cells transiently transfected with the deletions constructs shown in panel B. Analysis of the data reveals that deletions made in the stem result in increased exon 4 skipping. (D) Quantitation of the data in panel C.