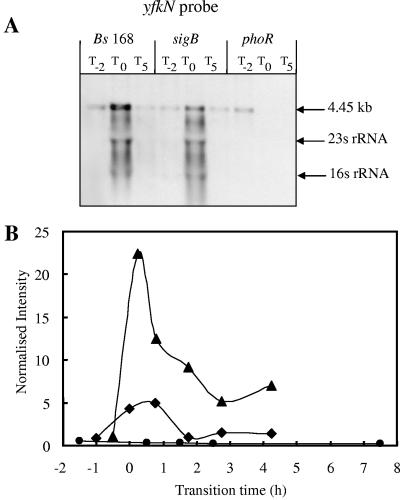
FIG. 4.
Northern blot analyses and transcriptional profiles for the yfkN gene. (A) RNA was isolated from wild-type B. subtilis strain 168 (Bs 168) and sigB- and phoR-null mutants. Bacteria were grown in LPM, and samples were taken 2 h before (T−2) and 0 h (T0) and 5 h (T5) after entry into the stationary growth phase, which was provoked by phosphate starvation. Five micrograms of RNA was applied per lane; after the filters were capillary blotted, they were hybridized to yfkN-specific riboprobes. Transcript size was determined by comparison with DIG-labeled RNA size markers (Roche Diagnostics, Mannheim, Germany). The arrows labeled 16S rRNA and 23S rRNA indicate the locations of these rRNA species that are known to trap smaller RNA species (1). (B) RNA was isolated from wild-type B. subtilis (168) (♦) and sigB-null (▴), and phoR-null (•) mutants grown in LPM. Samples were taken before, during, and after entry into the stationary growth phase, which was provoked by phosphate starvation. Total RNA was isolated and used as the template for reverse transcriptase incorporating radiolabeled [33P]dATP, which was hybridized to a whole-genome macroarray (Sigma-Genosys, The Woodlands, Tex). Normalization and quantification were performed as described in Materials and Methods.