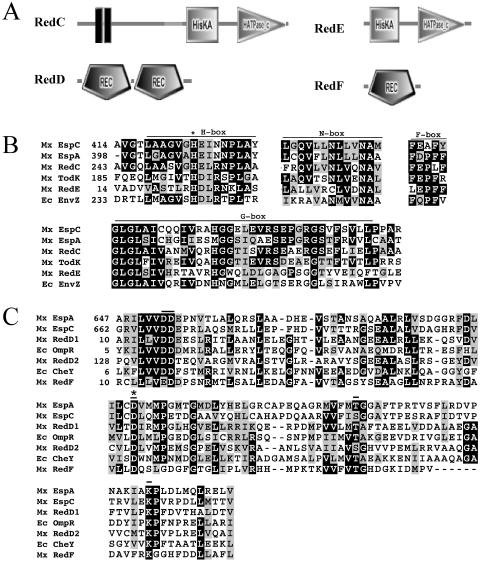
FIG. 2.
Domain architecture and sequence alignments of histidine kinase and receiver domains. A) Arrangement of signal transduction domains of the two-component signal transduction proteins predicted by SMART. Histidine kinase (HisKA) and ATP-binding (HATPase_c) domains are depicted in RedC and RedE. Receiver (REC) domains are depicted in RedD and RedF. RedC was modified to add an additional transmembrane domain predicted by transmembrane hidden Markov model (16). B) Sequence alignments of RedC and RedD compared to several histidine kinases shown to modulate developmental progression in M. xanthus (EspC [9], EspA [3], TodK [12]) and to canonical histidine kinase EnvZ from E. coli (Ec EnvZ) (11). Conserved regions within the HisKA (H box) and HATPase_c (N, D, F, and G boxes) domains are shown. An asterisk denotes the conserved histidine residue which is the site of autophosphorylation in EnvZ (8). C) Receiver domains identified in RedD and RedF were aligned with those from the hybrid kinases EspA and EspC and canonical response regulator proteins OmpR and CheY from E. coli. Important functional residues are indicated by bars. An asterisk denotes the conserved aspartate residue which is the site of autophosphorylation in OmpR and CheY (19). Mx, M. xanthus; Ec, E. coli.