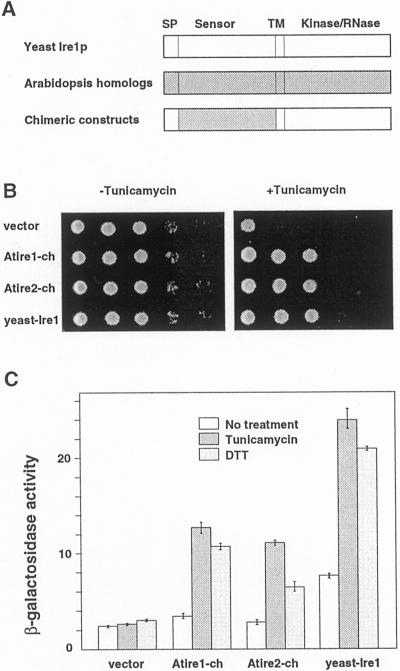
Figure 6.
A through C, Complementation of yeast ire1 deletion mutant with chimeric constructs of yeast Ire1p and Arabidopsis Ire1 homologs. A, Schematic view of the chimeric constructs used for yeast complementation. Chimeric constructs consisted of the sensor domains of AtIre1-1 or AtIre1-2, and other parts (signal peptide and C-terminal half) of yeast Ire1p. B, Growth of Δire1 strains of yeast complemented with chimeric constructs in the absence or presence of tunicamycin. Δire1 containing vector only (vector), chimeric constructs for AtIre1-1 (Atire1-ch) and AtIre1-2 (Atire2-ch), and yeast IRE1 were grown on synthetic dextrose plates without tunicamycin (−Tunicamycin) and with tunicamycin (+Tunicamycin, final concentration at 0.2 μg mL−1). Approximately 1 × 105 cells were spotted on the left column of each plate. Series of one-tenth dilution of cells were spotted on the right side. Yeast cells were grown for 3 d at 30°C. C, Activity of β-galactosidase of yeast cells containing each construct. Cells were incubated at 30°C with 2 μg mL−1 tunicamycin for 4 h, or 1 mm dithiothreitol for 2 h, and their β-galactosidase activity was measured.