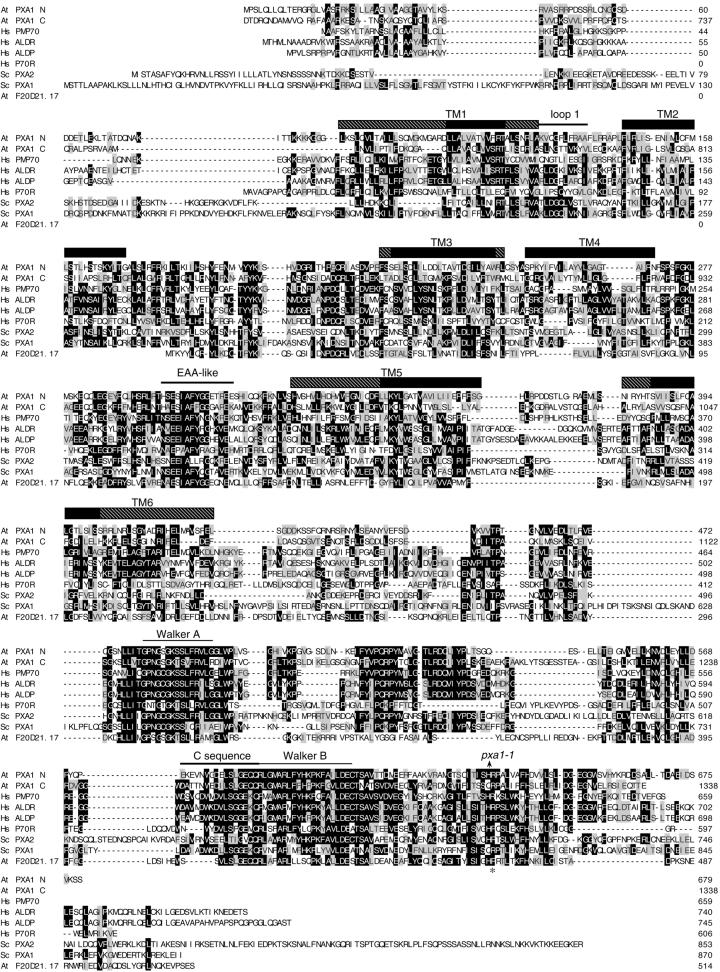
Figure 6.
Alignment of PXA1 and its homologs. Alignment of PXA1 with the PMP70 (GenBank accession no. XP_010507, 45% identical to Arabidopsis PXA1), ALDR (NP_005155, 42% identical), ALDP (XP_010174, 42% identical), and P70R (NP_064731, 36% identical) proteins from human (Hs); the Pxa2p (NP_012733, 30% identical) and Pxa1p (NP_015178, 24% identical) proteins from yeast (Sc); and the F20D21.17 (AAD25615, 21% identical) protein from Arabidopsis (At). The PXA1 protein is divided into halves to show its homology with the hemitransporters; PXA1 N is the N terminus of the protein from amino acids 1 to 679; PXA C is the C terminus of the protein from amino acids 680 to 1,338. Sequences were aligned with the MegAlign program (DNASTAR, Inc., Madison, WI) using the ClustalW method. Amino acid residues identical in at least three of the sequences are boxed in black and similar amino acids are boxed in gray. Hyphens indicate gaps introduced to maximize alignment. The arrow above the alignment marks the position of the pxa1 splicing defect and conserved domains are indicated above the sequence. The 12 TM domains were predicted using homology to the human (Shani and Valle, 1998; Dubois-Dalcq et al., 1999) and yeast proteins (Shani and Valle, 1996; Swartzman et al., 1996) and by TM prediction programs TMAP (Perrson and Argos, 1994, 1996), SMART; (Schultz et al., 1998, 2000), and TMpred (Hofmann and Stoffel, 1993). TM domains are indicated by rectangles above the sequence; regions of high certainty are indicated by black boxes; regions of lower certainty are indicated by hatched boxes. An asterisk marks an Arg residue in ALDP that is a site of recurrent mutations in X-ALD patients (Dubois-Dalcq et al., 1999), which coincides with the first amino acid affected in the pxa1 mutant.