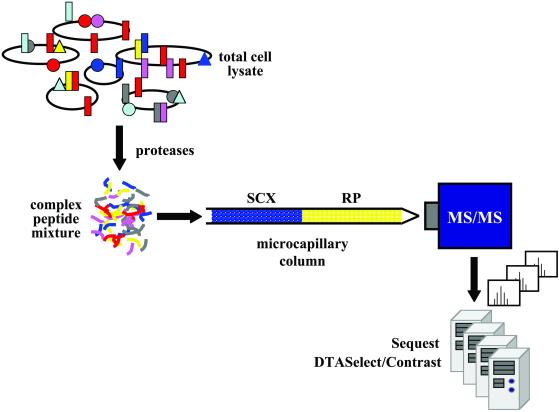
Figure 1.
Multidimensional Protein Identification Technology (MudPIT). The complex mixture of proteins present in a whole cell lysate is fragmented first with lysine-specific endoproteinase lysC in the presence of 8 M urea and then with immobilized trypsin, after dilution to 2 M urea, generating a highly complex mixture. The peptides are collected on a strong cation exchange (SCX) column that is positioned immediately upstream of a reverse-phase (RP) column. Successive peptide fractions are released, depending on their isoelectric point, with salt steps of increasing concentration at low organic solvent concentrations and captured by the second-dimension reverse-phase column. The reverse-phase column is eluted with a gentle gradient of increasing organic solvent concentration between each salt step to displace the peptides, depending on their hydrophobicity, into the mass spectrometer. The ion-trap mass spectrometer (LCQ-DECA, ThermoFinnigan, San Jose, CA) employs data-dependent acquisition software to limit the time spent sequencing any particular peptide, so that as many different peptides as possible are sequenced, regardless of their abundance. sequest software correlates experimental sequence with genomic data (courtesy of Christine Wu, The Scripps Institute, La Jolla, CA).