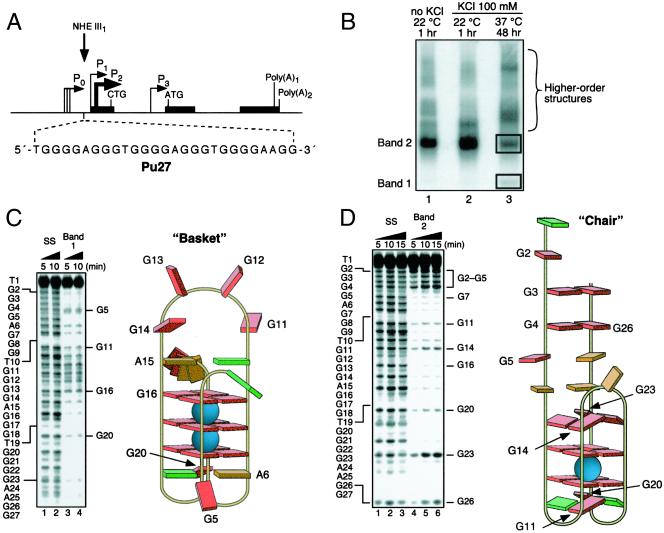
Figure 1.
Determination of the structures of the unimolecular G-quadruplexes formed after incubation of the Pu 27 strand (Pu27) in 100 mM KCl for 48 h at 37°C. (A) Promoter structure of the c-MYC gene; shown in Inset is the 27-mer sequence of the purine-rich strand upstream of the P1 promoter (3). (B) Nondenaturing gel analysis (15% polyacrylamide/12.5 mM KCl/NaCl, 4°C, 16 h) of Pu 27 preincubated under the conditions specified in the figure at a strand concentration of ≈25 μM. (C) DMS footprinting of band 1 in B. (Left) DMS treatment of the denatured Pu27 (lanes 1 and 2) and the isolated band (lanes 3 and 4). The Pu27 base sequence is shown to the left. (Right) Proposed structure based upon the footprinting pattern. Guanines showing DMS-induced cleavage are labeled in both Left and Right. Base colors: red, guanine; green, thymine; and orange, adenine. (D) As in C, but for band 2 in B.