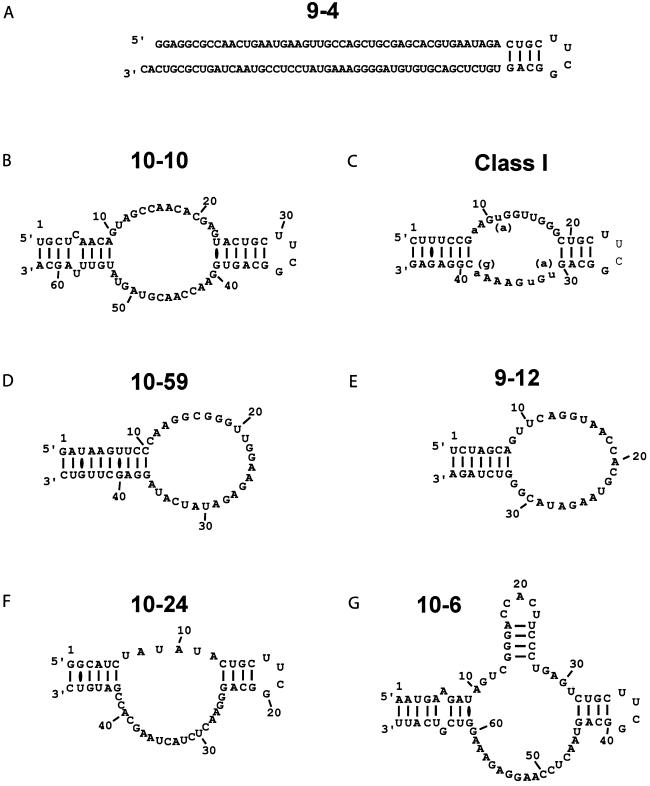
Figure 5.
Sequences of the highest-affinity RNA aptamers, in order of their measured binding constants (see Table 2), and their proposed secondary structures. The lengths were determined by end mapping, and the secondary structures based on the assumption that complementary termini form a duplex, and that the engineered hairpin is formed whenever present. G (10-6) has a potential 4-base stem within the binding loop, and A (9-4) has a number of possible structures, so no particular structure is presented. C, a Class I aptamer, is the best characterized structure, because 13 independent variants emerged from the selection. The variable positions in the binding loop (see Fig. 4) are denoted by lowercase letters.