Table 1.
Masses and sedimentation coefficients of HD exon 1, TRX-tag, and MW1 Fab
| Protein | Mass, kDa
|
s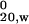 ,‡ S ,‡ S |
f/f0§ | |||
|---|---|---|---|---|---|---|
| Calculated* | MS† | FPLC† | AUC† | |||
| HD-25Q | 25.2 | 25.2 | 56.4 | 26.2 | 1.58 | 1.9 |
| HD-46Q | 27.9 | 27.9 | 61.0 | 27.2 | 1.66 | 2.0 |
| TRX-tag | 14.8 | 14.8 | 19.8 | 15.2 | 1.56 | 1.4 |
| MW1 Fab | 47‖ | 47.5 | 43.8 | 37.2 | 3.30 | 1.4 |
| Typical values | ||||||
| Elongated | >1.6 | |||||
| Globular | ≈1.2–1.3 | |||||
Calculated mass for protein expressed in bacteria, assuming cleavage of N-terminal Met.
Masses determined by mass spectrometry (MS) or sedimentation velocity analytical ultracentrifugation (AUC), or estimated from gel filtration chromatography (FPLC). Although fusion proteins with normal or expanded poly(Gln) migrate in gel filtration with high apparent molecular weights, they are monomeric as shown by AUC.
Three concentrations each of HD-25Q, HD-46Q, or TRX-tag were analyzed by sedimentation velocity and apparent sedimentation coefficients (s*) were extrapolated to infinite dilution and adjusted to standard conditions of 20°C in water (s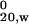 ). A single concentration of MW1 Fab was analyzed and correction of s* to infinite dilution was estimated with the program sednterp (33).
). A single concentration of MW1 Fab was analyzed and correction of s* to infinite dilution was estimated with the program sednterp (33).
Ratio of the observed frictional coefficient (f) and that calculated for an anhydrous sphere of equivalent radius (f0), calculated according to the Teller method with sednterp (33).
Mean of calculated masses from IgG2b/κ Fab sequences in the Protein Data Bank, PDB codes 1IBG, 1FAI, and 1CGS.
