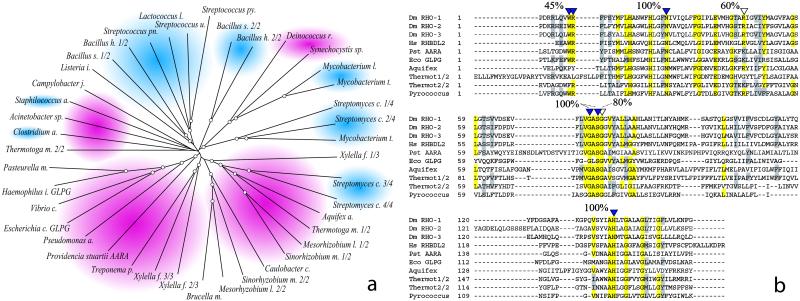
Fig 1.
RHOr sequences are widespread among bacteria. (a) Phylogenetic tree derived from the alignment of 41 bacterial RHOr. Each sequence is represented by the name of the bacterial genus from which it derives. To facilitate distinction of different species of the same genus, the first letter of the species name is also reported. Gram-negative groups are shaded in red, Gram-positive groups are shaded in blue. Tree nodes with a bootstrap support greater than 50% are encircled. With a few exceptions, Gram-negative and positive sequences group separately. In addition, within each group at least two subfamilies seem to resolve indicating perhaps an ancient event of gene duplication. (b) The RHO-domain of all of the proteins that have been shown able to activate SPI is aligned to the one of AarA and of a few representative prokaryotic sequences. Residues that are essential for RHO-1 function are indicated by a full arrowhead. Residues that, when mutated, decrease RHO-1 activity without abolishing it, are designated by empty arrowheads. A total of 41 bacterial RHOr where aligned as above, and the conservation of catalytic residues determined. The percent value reported above the alignment represents the approximate level of conservation of the key residues, a positive score was assigned if residues where found absolutely conserved (in yellow) or very similar (in gray). In AarA and GlpG all of the catalytic residues are absolutely conserved.