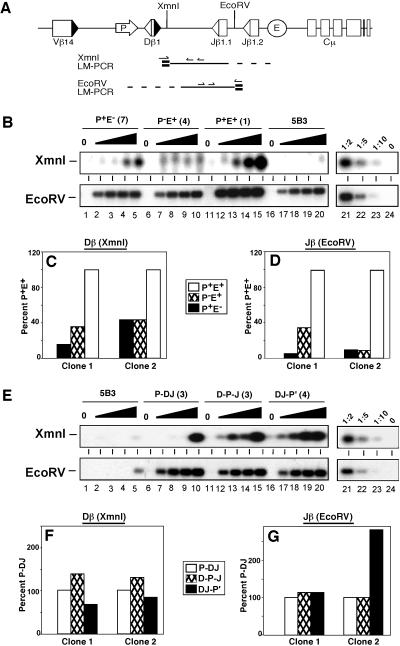
Fig 3.
Restriction endonuclease sensitivity is independent of promoter location. (A) Diagram of LM-PCR strategies for XmnI or EcoRV cleavage products. BW-1/2 linkers are shown in bold, and TCRβ-specific primers are indicated by arrows. (B and E) Levels of RE cleavage products within modified substrates. Nuclei from 5B3 clones containing the indicated TCRβ substrate or untransfected 5B3 cells were incubated with XmnI (0, 0.5, 1, 5, and 10 units) or EcoRV (at 0, 0.1, 0.5, 1, and 5 units). Genomic DNA from treated nuclei was subjected to nested LM-PCR for cleavage products (A). The linearity of each assay was confirmed by serial dilutions (lanes 21–24) of the maximally digested P+E+ sample (lane 15). (C, D, F, and G) Quantification of RE sensitivity in TCRβ miniloci. LM-PCR signals for XmnI (5 units) or EcoRV (5 units) treatment were quantified by PhosphorImager analysis. Values were normalized to signals obtained with PCR assays for DNA content (Cλ), substrate copy number (Jβ1.1), and cleavage of accessible loci (Cκ for XmnI or c-myc for EcoRV). Normalized values are shown relative to data for P+E+ (C and E) or P-DJ (D and F).