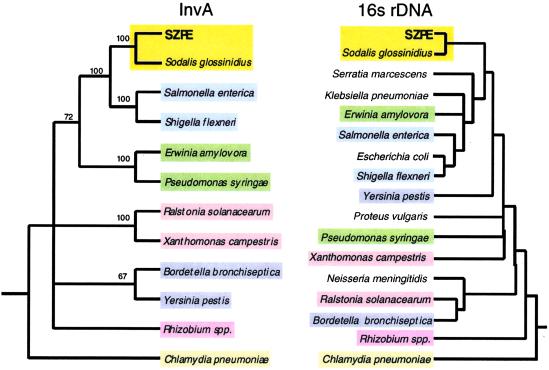
Fig 1.
Phylogenetic trees based on the InvA and 16S rDNA sequences from SZPE, S. glossinidius, and selected plant and animal pathogens. For the InvA tree, only clades supported by bootstrap values greater than 60% are shown as resolved. Accession numbers of InvA sequences are as follows: SZPE (AF467290), S. glossinidius (AF306649), Yersinis pestis (NC001972), S. enterica (U43239), S. flexneri (NC002698), P. syringae (AAG33877), Ralstonia solanacearum (P35656), Chlamydia pneumoniae (NC000922), Erwinia amylovora (X99768), Rhizobium sp. NGR234 (P55726), Xanthomonas campestris (P80150). The invA homolog from Bordetella bronchiseptica was obtained from a dataset produced by the Bordetella bronchiseptica Sequencing Group at the Sanger Center, and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/bb/.