In eukaryotic cells, intracellular trafficking is essential for the maintenance of the membrane integrity of organelles. It involves transport of molecules (such as proteins and lipids) by vesicles from a donor compartment and to a receiving (or target) compartment. Once the vesicle is docked properly on the target membrane, fusion occurs to complete the delivery of the cargo. The fusion step is believed to be mediated by a set of membrane proteins called SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins (1). One SNARE protein (called v-SNARE) resides exclusively on the transport vesicle, whereas two or more SNARE proteins (called t-SNAREs) usually reside on the target membrane. V- and t-SNAREs recognize each other and assemble into biochemically stable trans-SNARE complexes, which bring the vesicle close to the target membrane and mediate vesicle fusion.
Perhaps the SNARE proteins not only drive membrane fusion but they also determine the specificity of vesicle delivery.
One intriguing question is what determines the specificity of vesicle targeting, docking, and fusion. Examination of the subcellular distribution of SNARE proteins reveals that different SNARE proteins are localized to distinct membrane compartments (2). This observation raises the possibility that perhaps the SNARE proteins not only drive membrane fusion but that they also determine the specificity of vesicle delivery by selective assembly of SNARE complexes. The idea that SNARE proteins mediate vesicle fusion and define trafficking specificity constitutes the core of the SNARE hypothesis postulated by Rothman and coworkers (1, 3). Although the role of SNAREs in vesicle fusion has been generally accepted, the issue of vesicle docking and cargo delivery specificity remains a subject of intense debate. In this issue of PNAS, Schwarz and colleagues (4) demonstrate that two v-SNAREs with different tissue localizations and functions are able to substitute for each other to maintain cell viability and synaptic vesicle exocytosis in the fruit fly Drosophila. Furthermore, they show that homologous v-SNAREs from rat function equally well in place of the fly proteins. These results offer a different view of the specificity of vesicle trafficking conferred by the SNARE proteins.
The most compelling evidence to date supporting a critical role for SNARE proteins in determining vesicle fusion specificity comes from systematic studies of a large number of SNARE proteins encoded in the yeast genome. Rothman and coworkers (5) used an in vitro method to study vesicle fusion by measuring lipid content mixing from two separate liposomal vesicles, each one reconstituted with v-SNARE and t-SNAREs, respectively. In a series of recent studies (6–9), these investigators examined whether different pairs of SNARE proteins could mediate vesicle fusion in vitro. Remarkably, their results revealed that only a few pairs of SNARE proteins among hundreds of possible combinations could mediate vesicle fusion, albeit at a nonphysiological rate. Most other SNARE pairs resulted in either an inefficient fusion or no fusion at all. The authors concluded that specific pairing of cognate SNARE proteins (i.e., SNAREs found in the same membrane compartment) provides the inherent molecular mechanism for vesicle docking and compartmental specificity.
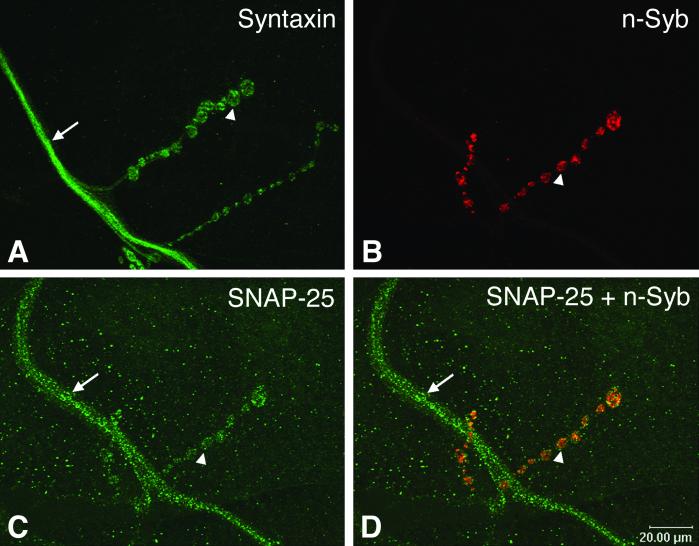
These new findings offer an attractively unified mechanism for vesicle fusion and trafficking specificity, which may explain why a particular vesicle fuses with the plasma membrane instead of the lysosome or any other intracellular organelles. However, the idea that compartmental specificity is achieved through specific SNAREs is not without its caveats, and alternative views merit consideration. First, biochemical studies have shown that SNARE proteins that normally do not reside in the same membrane compartment readily form stable complexes (10, 11), although some complexes were later shown to be ineffective in mediating fusion in cracked PC12 cells (12). Second, it remains unclear whether the in vitro fusion assay closely reflects vesicle fusion in vivo, where a large number of proteins exist to modulate the formation of trans-SNARE complexes. For example, the cytosolic proteins complexin and m/unc-13 have been shown to promote the formation of SNARE complex and vesicle fusion (13–17). Thus, the possibility remains that some, if not all, of the SNARE complexes that failed to mediate the aforementioned liposomal fusion may drive fusion efficiently when the appropriate in vivo cofactors are present. Third, it remains to be determined whether the diversity of subcellular localization of SNARE proteins is sufficient to determine compartmental specificity. One of the puzzling observations on the subcellular localization of SNARE proteins is that they do not always restrict their locations to the site of action. As shown in Fig. 1, the Drosophila t-SNAREs, syntaxin and SNAP-25, are localized not only to the synaptic terminal, but also on the entire axonal membrane (18, 19). However, the v-SNARE synaptobrevin is found exclusively with synaptic vesicles within the synapse (20). How do synaptic vesicles know to bypass the t-SNAREs along the axon and localize to the nerve terminal? Given this apparently paradoxical localization of SNARE proteins, it is reasonable to speculate that SNARE proteins are not involved in vesicle targeting and docking. This notion is supported by genetic studies in Drosophila (18, 20–22), C. elegans (23), and mouse (24, 25), and by toxin injection studies in the squid giant axon (26). These studies have demonstrated that perturbation of SNARE proteins, whether it is a v-SNARE or a t-SNARE, does not affect synaptic vesicle targeting and docking. In fact, the lack of spatial restriction of SNARE proteins may be a common feature also found outside the nervous system. For example, the Drosophila t-SNARE syntaxin is known to be required for vesicle fusion in all tissues (18, 27). Instead of being restricted to the sites of active exocytosis, the yeast tSNARE sec9 (a SNAP-25 homolog) is distributed over the entire plasma membrane (28). Finally, the yeast v-SNARE Vti1p has also been shown to interact with multiple t-SNAREs and functions in at least three trafficking pathways (29).
Figure 1.
Immunocytochemical localization of t- and v-SNARE proteins at wild-type Drosophila larval neuromuscular junction. (A, C, and D) The t-SNARE proteins, syntaxin (green color) and SNAP-25 (green color), are localized to both axons and synaptic boutons. (B and D) The v-SNARE protein neuronal synaptobrevin (red color) is restricted to and enriched in synaptic boutons. B and C are from the same neuromuscular junction preparation, whereas D is an overlap of B and C. Synaptic termini (called synaptic boutons) are recognized by their unique beads-on-a-string morphology (one of the boutons is indicated by an arrowhead in each panel). The axon is indicated by an arrow. The muscle on which the synapse forms is invisible here. Based on these staining patterns, it is unlikely that pairing between these cognate v- and t-SNAREs could target and dock synaptic vesicles specifically to the synaptic terminal but not the axon. The monoclonal syntaxin antibody (8C3) developed by S. Benzer (California Institute of Technology, Pasadena, CA; ref. 40) was obtained from the Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA; a rabbit SNAP-25 polyclonal antibody was a gift from D. Deitcher (Cornell University, Ithaca, NY; ref. 19); and a rat n-Syb polyclonal antibody (R29) was a gift from H. Bellen (Baylor College of Medicine, Houston; ref. 41).
Now Schwarz and coworkers (4) have provided a direct test of the role of SNARE proteins in the specificity of vesicle fusion in an in vivo system. In Drosophila, there are only two characterized v-SNAREs, a ubiquitous synaptobrevin (called syb) and a neuron-specific synaptobrevin (called n-syb). Deletion of n-syb reduces spontaneous vesicle fusion and completely blocks action potential-evoked vesicle exocytosis (20). These results suggest that normally syb does not play a redundant role in synaptic transmission. Similarly, n-syb does not normally function in the membrane compartments reserved for syb, because n-syb is not expressed outside the nervous system. Syb and n-Syb seem to overlap in the eye (see below), but there is no functional overlap between them, suggesting that they are in separate membrane compartments. Despite their distinct compartmental functions, bacterium-expressed recombinant syb and n-syb form stable SNARE complexes with the plasma membrane t-SNAREs syntaxin and SNAP-25 in vitro. Are these two v-SNAREs functionally interchangeable in vivo? By using a new genetic method recently developed in the Schwarz laboratory (30), the authors generated mutations of these two synaptobrevins but restricted the mutation to the compound eye only. When syb was removed from the eye precursor cells, the eye failed to develop. In contrast, removal of n-syb did not have a significant effect on eye formation, but it interrupted synaptic transmission from photoreceptors to downstream neurons. They then asked whether the mutant phenotypes caused by deleting one type of synaptobrevins could be rescued by expressing wild type copies of the other synaptobrevin. Their results show that each synaptobrevin can functionally replace the other. Furthermore, homologs of syb (cellubrevin) and n-syb (VAMP2) from rat also function equally well in genetic rescue of syb mutations. To confirm further the role of syb in rescuing synaptic transmission, these investigators used a heatshock promoter to express syb in the n-syb mutant and achieved partial rescue of evoked-transmitter release at the neuromuscular junction (NMJ) synapse. It is important to note that syb expression rescued synaptic transmission to the same degree as that by n-syb expression. The results from these elegant genetic experiments argue that SNARE proteins may not be sufficient for vesicle trafficking and fusion specificity.
Could this v-SNARE swapping result be an exception rather than the rule? It is interesting to note that other genetic studies are consistent with the findings by the Schwarz group. In yeast, two yeast syntaxin homologs, Pep12p and Vam3p, function as endosomal and vacuolar t-SNAREs, respectively. Mutations of Pep12p specifically affect vesicle trafficking from the late Golgi to the endosome, whereas mutations in Vam3p only affect vesicle fusion with the vacuole. However, Pep12p and Vam3p can functionally rescue each other's mutant phenotype when they are overexpressed (31, 32). Increasing evidence suggests that there is also a functional redundancy of SNARE proteins in vesicle trafficking regulated by endogenous transcription regulation. The yeast Sec22p is required for fusion of endoplasmic reticulum (ER)-derived vesicles with early Golgi membranes, whereas Ykt6p is a SNARE protein functioning at late stages of the secretory pathway. In the absence of Sec22p, Ykt6p is up-regulated and functionally supports the ER-Golgi pathway (33). Similarly, depletion of SNAP-25 has no impact on synaptic transmission in Drosophila larval NMJs, likely because of compensation by another t-SNARE, SNAP-24 (34). These genetic studies collectively suggest that SNARE pairing is insufficient to confer vesicle-fusion specificity.
The SNARE hypothesis has provided an elegant conceptual framework for cell biological studies of intracellular trafficking. Driven by this major force, we have experienced a decade of unprecedented advances toward understanding vesicle fusion. However, it is also clear that some tenets of the SNARE hypothesis have been challenged and perfected by recent studies (2, 6). Although the SNARE proteins may take central stage in the years to come, it will be beneficial also to focus on several other proteins that have emerged recently as potentially key players in trafficking and fusion specificity (35, 36). The exocyst sec6/8 complex has been implicated as putative tethering proteins (37), whereas the synaptic vesicle protein synaptotagmin has been shown to stabilize synaptic vesicle docking (38). In some cases, molecules that are least suspected may turn out to be gold mines. For example, a recent genetic study revealed that synaptic vesicles are found along the axon in mouse neuromuscular junctions deficient in the cell adhesion molecule NCAM (39). Interestingly, these “lost” vesicles are fully capable of releasing transmitter through exocytosis. Future studies of non-SNARE proteins may lead us to a better understanding of mechanisms of vesicle docking and the specificity of vesicle fusion.
Footnotes
See companion article on page 13867.
References
- 1.Sollner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y A, Scheller R H. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 3.Rothman J E. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya S, Stewart B A, Niemeyer B A, Burgess R W, McCabe B D, Lin P, Boulianne G, O'Kane C J, Schwarz T L. Proc Natl Acad Sci USA. 2002;99:13867–13872. doi: 10.1073/pnas.202335999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber T, Zemelman B V, McNew J A, Westermann B, Gmachl M, Parlati F, Sollner T H, Rothman J E. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 6.McNew J A, Parlati F, Fukuda R, Johnston R J, Paz K, Paumet F, Sollner T H, Rothman J E. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 7.Parlati F, McNew J A, Fukuda R, Miller R, Sollner T H, Rothman J E. Nature. 2000;407:194–198. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda R, McNew J A, Weber T, Parlati F, Engel T, Nickel W, Rothman J E, Sollner T H. Nature. 2000;407:198–202. doi: 10.1038/35025084. [DOI] [PubMed] [Google Scholar]
- 9.Parlati F, Varlamov O, Paz K, McNew J A, Hurtado D, Sollner T H, Rothman J E. Proc Natl Acad Sci USA. 2002;99:5424–5429. doi: 10.1073/pnas.082100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang B, Gonzalez L, Jr, Prekeris R, Steegmaier M, Advani R J, Scheller R H. J Biol Chem. 1999;274:5649–5653. doi: 10.1074/jbc.274.9.5649. [DOI] [PubMed] [Google Scholar]
- 11.Fasshauer D, Antonin W, Margittai M, Pabst S, Jahn R. J Biol Chem. 1999;274:15440–15446. doi: 10.1074/jbc.274.22.15440. [DOI] [PubMed] [Google Scholar]
- 12.Scales S J, Chen Y A, Yoo B Y, Patel S M, Doung Y C, Scheller R H. Neuron. 2000;26:457–464. doi: 10.1016/s0896-6273(00)81177-0. [DOI] [PubMed] [Google Scholar]
- 13.Reim K, Mansour M, Varoqueaux F, McMahon H T, Sudhof T C, Brose N, Rosenmund C. Cell. 2001;104:71–81. doi: 10.1016/s0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 14.Tokumaru H, Umayahara K, Pellegrini L L, Ishizuka T, Saisu H, Betz H, Augustine G J, Abe T. Cell. 2001;104:421–432. doi: 10.1016/s0092-8674(01)00229-x. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Tomchick D R, Kovrigin E, Arac D, Machius M, Sudhof T C, Rizo J. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 16.Richmond J E, Weimer R M, Jorgensen E M. Nature. 2001;412:338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brose N, Rosenmund C, Rettig J. Curr Opin Neurobiol. 2000;10:303–311. doi: 10.1016/s0959-4388(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 18.Schulze K L, Broadie K, Perin M S, Bellen H J. Cell. 1995;80:311–320. doi: 10.1016/0092-8674(95)90414-x. [DOI] [PubMed] [Google Scholar]
- 19.Rao S S, Stewart B A, Rivlin P K, Vilinsky I, Watson B O, Lang C, Boulianne G, Salpeter M M, Deitcher D L. EMBO J. 2001;20:6761–6771. doi: 10.1093/emboj/20.23.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deitcher D L, Ueda A, Stewart B A, Burgess R W, Kidokoro Y, Schwarz T L. J Neurosci. 1998;718:2028–2039. doi: 10.1523/JNEUROSCI.18-06-02028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broadie K, Prokop A, Bellen H J, O'Kane C J, Schulze K L, Sweeney S T. Neuron. 1995;15:663–673. doi: 10.1016/0896-6273(95)90154-x. [DOI] [PubMed] [Google Scholar]
- 22.Sweeney S T, Broadie K, Keane J, Niemann H, O'Kane C J. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 23.Nonet M L, Saifee O, Zhao H, Rand J B, Wei L. J Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoch S, Deak F, Konigstorfer A, Mozhayeva M, Sara Y, Sudhof T C, Kavalali E T. Science. 2001;294:1117–1122. doi: 10.1126/science.1064335. [DOI] [PubMed] [Google Scholar]
- 25.Washbourne P, Thompson P M, Carta M, Costa E T, Mathews J R, Lopez-Bendito G, Molnar Z, Becher M W, Valenzuela C F, Partridge L D, Wilson M C. Nat Neurosci. 2002;5:19–26. doi: 10.1038/nn783. [DOI] [PubMed] [Google Scholar]
- 26.Hunt J M, Bommert K, Charlton M P, Kistner A, Habermann E, Augustine G J, Betz H. Neuron. 1994;12:1269–1279. doi: 10.1016/0896-6273(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 27.Burgess R W, Deitcher D L, Schwarz T L. J Cell Biol. 1997;138:861–875. doi: 10.1083/jcb.138.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brennwald P, Kearns B, Champion K, Keränen S, Bankaitis V, Novick P. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 29.Fischer von Mollard G, Stevens T H. Mol Biol Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stowers R S, Schwarz T L. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darsow T, Rieder S E, Emr S D. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotte M, Gallwitz D. FEBS Lett. 1997;411:48–52. doi: 10.1016/s0014-5793(97)00575-9. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Barlowe C. Mol Biol Cell. 2002;13:3314–3324. doi: 10.1091/mbc.E02-04-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilinsky I, Stewart B A, Drummond J, Robinson I, Deitcher D L. Genetics. 2002;162:259–271. doi: 10.1093/genetics/162.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whyte J R, Munro S. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 36.Bajjalieh S M. Curr Opin Neurobiol. 1999;9:321–328. doi: 10.1016/s0959-4388(99)80047-6. [DOI] [PubMed] [Google Scholar]
- 37.Finger F P, Novick P. J Cell Biol. 1998;142:609–612. doi: 10.1083/jcb.142.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reist N E, Buchanan J, Li J, DiAntonio A, Buxton E M, Schwarz T L. J Neurosci. 1998;18:7662–7673. doi: 10.1523/JNEUROSCI.18-19-07662.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polo-Parada L, Bose C M, Landmesser L T. Neuron. 2001;32:815–828. doi: 10.1016/s0896-6273(01)00521-9. [DOI] [PubMed] [Google Scholar]
- 40.Fujita S C, Zipursky S L, Benzer S, Ferrus A, Shotwell S L. Proc Natl Acad Sci USA. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu M N, Fergestad T, Lloyd T E, He Y, Broadie K, Bellen H J. Neuron. 1999;23:593–605. doi: 10.1016/s0896-6273(00)80811-9. [DOI] [PubMed] [Google Scholar]