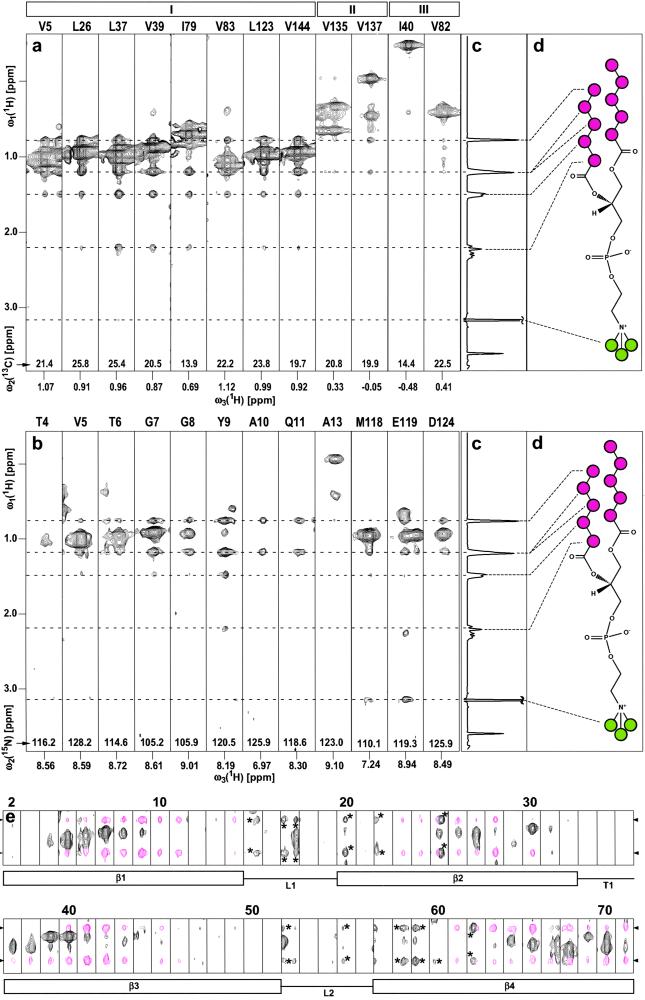
Figure 1.
(a) Selection of ω1(1H)/ω3(1H) strips from an 800-MHz 3D 13C-resolved [1H,1H]-NOESY spectrum measured with a sample of [u-2H,13C,15N/L,V,Iδ1-13CH3]-OmpX (23) in protonated DHPC micelles at 30°C (see Materials and Methods for details of the experimental set-up). The strips were taken at the 13C chemical shifts of the methyl groups for the residues indicated at the top, and they are centered about the methyl proton chemical shifts. The roman numbers I–III at the top denote different locations of the selectively protonated Val, Leu, and Ile(δ1) methyl groups relative to the surface of the NMR structure of OmpX/DHPC (see text). (b) Selection of ω1(1H)/ω3(1H) strips from an 800-MHz 3D 15N-resolved [1H,1H]-NOESY spectrum measured at 30°C with the same sample as in a. The strips were taken at the 15N chemical shifts of the residues indicated at the top and are centered about the respective amide proton chemical shifts. (c) One-dimensional 1H NMR spectrum of DHPC, measured with the same sample and the same experimental conditions as the spectra in a and b. (d) Chemical structure of DHPC. The CHn moieties of interest in this study are color-coded, with magenta circles indicating the CHn groups of the hydrophobic tails, and green circles identifying the polar head methyls of DHPC. The positions of the signals arising from the hydrophobic end (−CH3, δ = 0.78 ppm; penultimate and antepenultimate −CH2−, δ = 1.22 ppm; remaining −CH2− at δ = 1.50 and 2.21 ppm) and the choline N+-bound methyls (δ = 3.16 ppm) are marked with broken lines. (e) ω1(1H)/ω3(1H) strips for the polypeptide segment of residues 2–71 (same data set as in b), showing NOE cross peaks between backbone amide protons of the protein and the hydrophobic end groups of the DHPC. The chemical shifts of the DHPC protons shown here are indicated with arrowheads on the left and right. The NOE cross peaks of interest are colored magenta, and peaks labeled with asterisks represent tails of signals assigned in neighboring planes along ω2(15N). Below the strips, the secondary structure elements in this polypeptide segment are indicated, with β, L, and T standing for β-strand, loop, and turn, respectively.