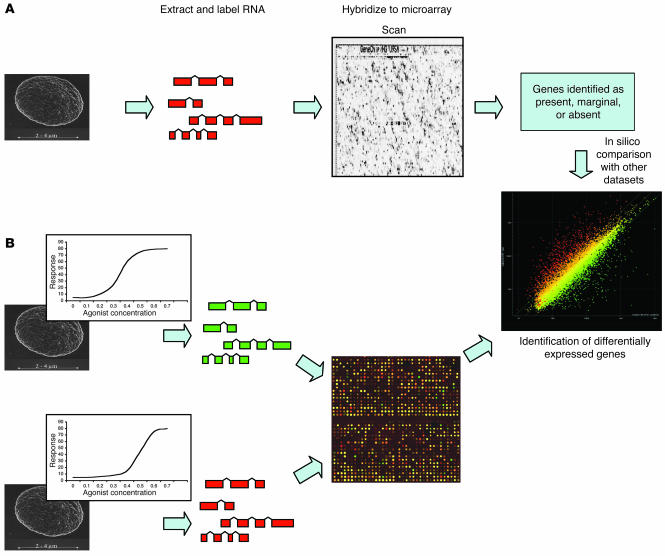
Figure 1.
Platelet transcript profiling by microarray. Generally, 1 of 2 approaches can be used to identify expressed transcripts in platelets. (A) With single-channel oligonucleotide arrays, mRNA is isolated from platelets, labeled, and hybridized to the microarray. The array is then processed and scanned, and genes are identified as “present,” “marginal,” or “absent.” Comparisons between samples can then be made in silico to identify differentially expressed genes. (B) In a 2-channel experiment, RNA from 2 individuals is isolated, each sample is labeled with a different fluorescent dye, and the 2 are compared directly on a microarray. In this case, the individuals have a different dose-response curve for a platelet agonist. Differentially expressed genes can then be directly identified.