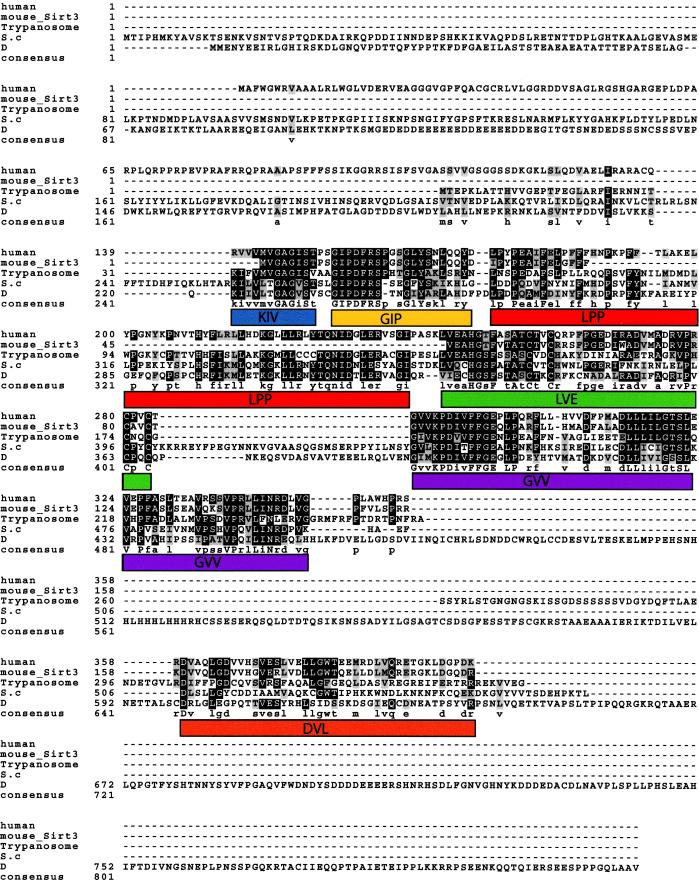
Figure 1.
Multiple sequence alignment of the mouse and human SIRT3 and trypanosoma, yeast (S.c), and Drosophila (D) SIR2 proteins. Identities are shaded black, and conserved substitutions are shown in gray. The cutoff for inclusion in the consensus is a 50% match in all the aligned proteins at a given position. Note the relative loss in conservation at the first 142 aa of SIRT3. The overall similarity between the mouse and the human gene is the best, understandably because of a more recent divergence in evolution, compared with the other organisms included in this analysis. Deduced motifs are represented by colored boxes and are given names derived from the first three letters shared from the consensus in all the proteins or, if conserved, in at least three of the five sequences. The motifs are: KIV, blue; GIP, yellow; LPP, red; LVE, green; GVV, purple; and DVL, orange. The KIV and GIP motifs span the conserved Sir2p core domain previously shown to be important in silencing (27).