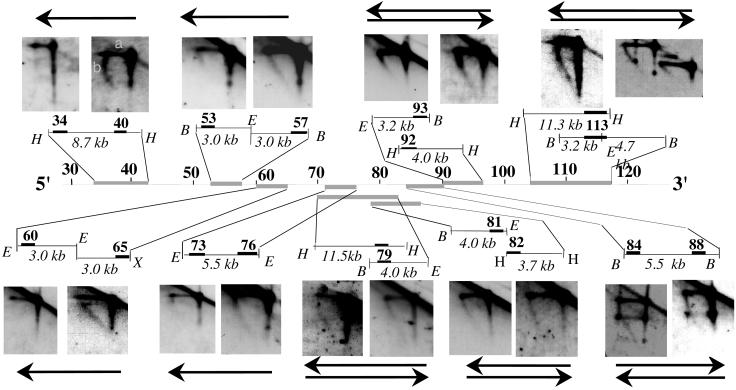
Figure 2.
Analysis of replication fork direction in the region downstream of the 3′ regulatory region by N/A 2D gel electrophoresis. The first dimension of the agarose gel is run in neutral conditions in which restriction enzyme-digested DNA molecules enriched in replication intermediates are separated according to their mass. The second dimension is run in alkaline conditions to separate parental and nascent strands. The direction of replication fork movement (indicated by arrow) is determined by hybridization to probes (solid rectangles) located at the 5′ or 3′ end of a specific restriction fragment (probes 199M11:34 and 40, 73 and 76, and 84 and 88); or from 5′ and 3′ ends of two adjacent restriction fragments (probes 199M11:53 and 57, 60 and 65, 81 and 82, and 92 and 93); or by using a single probe located at the ends of two overlapping fragments (probes 199M11:79 and 113). The size of the restriction fragment is indicated. Note that 199M11:34 and 199M11:117 were previously termed Cα-45 and Cα-100, respectively. Each segment (indicated by gray lines) was analyzed by two or more 2D gels with similar findings. The topmost restriction fragment and the topmost arrow (if there are two) always refer to the left of the two panels. The horizontal line (a) represents the parental strands of the replication intermediates, which run at a single position in the second dimension because they have a single molecular weight. The diagonal line (b) emanating from the parental strands represents nascent strands of different sizes, ranging from very small to as large as the parental strands. If replication forks move from 3′ to 5′, a probe located at the 3′ end of a fragment will detect nascent strands of sizes ranging from small to parental-sized, whereas probes located at the 5′ end will detect nascent strands only of a relatively large size. If forks move in both directions through a segment, then nascent strands of multiple sizes will be detected with probes for both 5′ and 3′ ends of this segment. Because of limitations in the resolution of N/A 2D gel electrophoresis, we cannot rule out the possibility that <10% of the forks progress from the opposite direction in the region upstream of 199M11:76. The spots present on the nascent strand arcs and the linear molecules on the autoradiogram probed with 199M11:84or :88 are caused by the accumulation of a single-stranded break in this segment, produced by EcoRI digestion, which is released during alkaline treatment in the 2nd dimension. No spots were detected on neutral/neutral (N/N) 2D gels. Therefore, this single-strand break does not appear to form any replication barrier. H, HindIII; E, EcoRI; B, BamHI; X, XbaI.