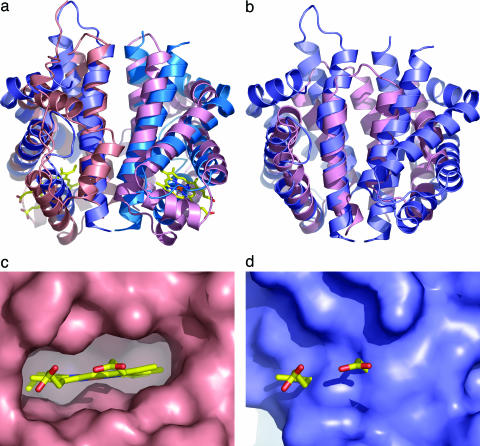
Fig. 2.
Comparison of N-RsbR with structural neighbors. (a) Protein cartoons of N-RsbR (shades of blue) and HemAT (shades of pink, PDB ID code 1OR4) after superimposition and in the same orientation as Fig. 1. The heme in HemAT is drawn as a “stick” figure. Each protein dimerizes through a four-helical bundle formed in each case by the C-terminal pairs of helices. Although there is little strict sequence homology between N-RsbR and HemAT residues in the dimer interface, hydrophobic amino acids in N-RsbR generally have hydrophobic equivalents in HemAT. The same trend can be observed for the polar and acidic amino acids. The Z-helix of HemAT can be seen running approximately between 1 o'clock and 7 o'clock in the figure in a groove, the floor of which comprises helices G and H, with walls of the loops between helices E and F to the left and the C–D corner to the right. (b) As a, but the structures of N-RsbR (blue) and the smaller N-RsbU (pink) are superimposed, and there is no equivalent in N-RsbU to the A–B helices of N-RsbR. (c) Close-up view of the heme pocket of HemAT, with the molecular surface depicted in pink and the bound heme drawn as a stick model, colored according to atom type. Note the complementarity of protein surface and heme and that the protrusion of surface immediately below the iron (brown) is the imidazole side chain of the proximal H123. (d) The same view as c of the equivalent region of N-RsbR (blue), with the heme from HemAT displayed for reference. Here, the surface pocket that accommodates heme in HemAT is filled in N-RsbR by the side chains of residues L37, V41, I45, E48, L70, R73, A74, I77, L79, F83, L84, and L88.