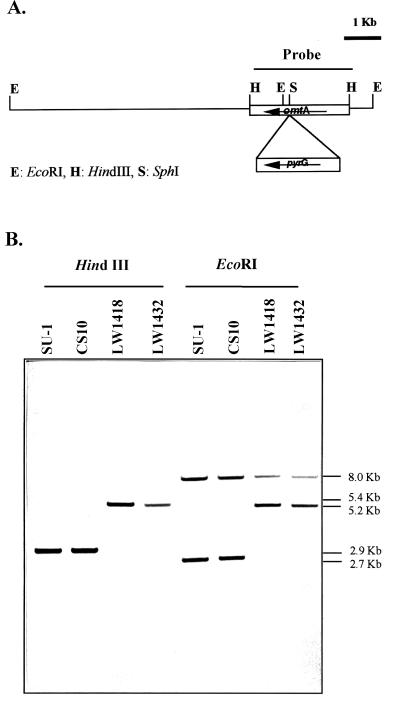
FIG. 2.
Southern hybridization analysis of omtA gene disruption strains. (A) Restriction endonuclease map of omtA locus in omtA disruption strains LW1418 and LW1432. (B) Southern hybridization analysis. Genomic DNAs isolated from LW1418 and LW1432, CS10 (parent strain) and SU1 (wild-type), were digested with restriction enzymes HindIII and EcoRI. DNAs were resolved and hybridized to a digoxigenin-labeled 2.9-kb HindIII omtA DNA fragment using standard methods. Strains with omtA disruption were predicted to contain a 5.4-kb HindIII fragment consisting of the 2.5-kb pyrG selectable marker inserted into the omtA locus. A 5.2-kb fragment in the omtA-disrupted strains was also predicted to replace the wild-type EcoRI fragment (2.7 kb). Numbers to the right of the blot represent the approximate size of the detected fragments in kilobase pairs.