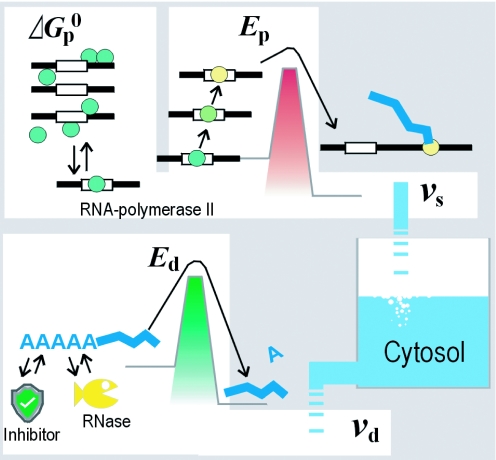
Figure 1.
Schematic representation of the proposed thermodynamic model. The accumulated amount of each transcript is in pseudo-equilibrium between influx and efflux. The velocity of synthesis for a transcript (vs) is determined according to the Gibbs free energy (for equilibrium between binding and dissociation of RNA polymerase II to the promoter) and activation energy Ep (energy barrier for the start of polymerase elongation). The velocity of degradation (vd) is determined by Ed (required to start hydrolysis). All energies are determined for interactions between special nucleotide sequences and the protein factors.