Abstract
Thymidine depletion is toxic to virtually all actively growing cells. The fundamental mechanism responsible for thymidineless death remains unknown. One event thought to be critical in causing the toxicity of thymidine depletion is a sharp rise in the ratio of dUTP to dTTP and subsequent incorporation of dUTP into DNA. Maneuvers to alter dUTP levels appear to alter the toxicity of thymidine depletion. However, loss of uracil-DNA-N-glycosylase activity does not appear to change the toxicity of thymidine deprivation significantly. This study proposes to define the role of uracil base excision repair (BER) in mediating thymidineless death. The toxicity of thymidine deprivation induced by the antifolate aminopterin was measured in a series of mutant Saccharomyces cerevisiae strains deficient in various steps in uracil-BER. Most mutants displayed modest changes in their sensitivity to aminopterin, with the exception of cells lacking the abasic endonuclease Apn1. apn1 mutants displayed a profound sensitivity to aminopterin that was relieved in an apn1 ung1 double mutant. Wild-type and apn1 mutants displayed similar levels of DNA damage and S-phase arrest during aminopterin treatment. A significant portion of cell killing occurred after removal of aminopterin in both wild-type and apn1 mutant cells. apn1 mutants showed a complete inability to re-initiate DNA replication following removal of aminopterin. These findings suggest recovery from arrest is a crucial step in determining the response to thymidine deprivation and that interruptions in uracil-BER increase the toxicity of thymidine deprivation by blocking re-initiation of replication rather than inciting global DNA damage. Inhibition of apurinic/apyrimidinic endonuclease may therefore be a reasonable approach to increase the efficacy of anticancer chemotherapies based on thymidine depletion.
INTRODUCTION
Depletion of thymidine is toxic to virtually all actively growing cells. Originally described by Barner and Cohen in 1954 (1), thymineless death has been reported in bacteria, yeast and mammalian cells (2). De novo synthesis of dTMP is carried out by the enzyme thymidylate synthase which converts dUMP to dTMP in a one carbon transfer to the 5′ position of dUMP. The one carbon group is donated by tetrahydrofolate which is regenerated by the enzyme dihydrofolate reductase. Decreased dTMP production can therefore be accomplished by either inhibiting thymidylate synthase or by inhibiting dihydrofolate reductase. The toxicity of thymidine depletion is the basis for chemotherapeutic agents including methotrexate (a dihydrofolate reductase inhibitor), fluorouracil and 5′-fluoro, 2′-deoxyuridine which are converted to a metabolite (FdUMP) capable of inhibiting thymidylate synthase.
The exact mechanism underlying thymidineless death remains elusive. Inhibition of thymidylate synthase not only decreases dTMP pools but also increases dUMP pools. Ingraham et al. (3) showed a 10 000 fold change in the ratio of dUMP to dTMP in cultured human lymphoblasts treated with methotrexate. Similar changes have been described by Tinkelenberg et al. (4) in a strain of Saccharomyces cerevisiae with a low activity allele of dUTPase. The rise in dUMP levels may result in a rise in dUTP levels, since dUMP is derived from dUTP. Most DNA polymerases have poor discrimination between dTTP and dUTP (5); therefore, a dramatic rise in the dUTP to dTTP ratio may result in significant incorporation of dUTP into DNA. Attempted repair of deoxyuridine residues from DNA without adequate dTTP available to complete the repair reaction has been suggested to create multiple single strand breaks, and eventually double strand breaks, in a so-called ‘futile cycle’ of repair (6). Indeed, single and double strand breaks do accumulate in thymidine deprived cells (7). In this model, loss of uracil glycosylase activity should decrease DNA breaks arising from attempted repair and thereby decrease the toxicity of thymidine depletion. However, loss of uracil glycosylase activity in some systems has no impact on the sensitivity of cells to thymidine deprivation (8).
Defining the role of uracil repair in mediating the toxicity of thymidine deprivation is challenged by the toxicity of excess amounts of uracil in DNA. dUTPase (dut) mutants of both yeast and Escherichia coli are inviable (9,10), potentially owing to high rates of dUTP incorporation into DNA. Thus, dut mutations and thymidine starvation both result in increased uracil laden DNA. Using a deletion mutation for dut, el-Hajj et al. (11) showed complete loss of dUTPase activity in a uracil glycosylase deficient strain of E.coli can lead to replacement of >90% of thymidine with uracil residues in DNA. The strain was not viable, implying that uracil laden DNA itself is toxic. In yeast, DUT1 is also an essential gene. Gadsden et al. (10) used a conditional allele of dUTPase to show loss of dUTPase activity leads to death. When dUTPase was inactivated in a uracil glycosylase deficient background, cells deficient in both dUTPase and uracil glycosylase actually lost viability more rapidly than isogenic cells lacking only dUTPase. These data suggest the presence of uracil in DNA may be more toxic than the attempted repair.
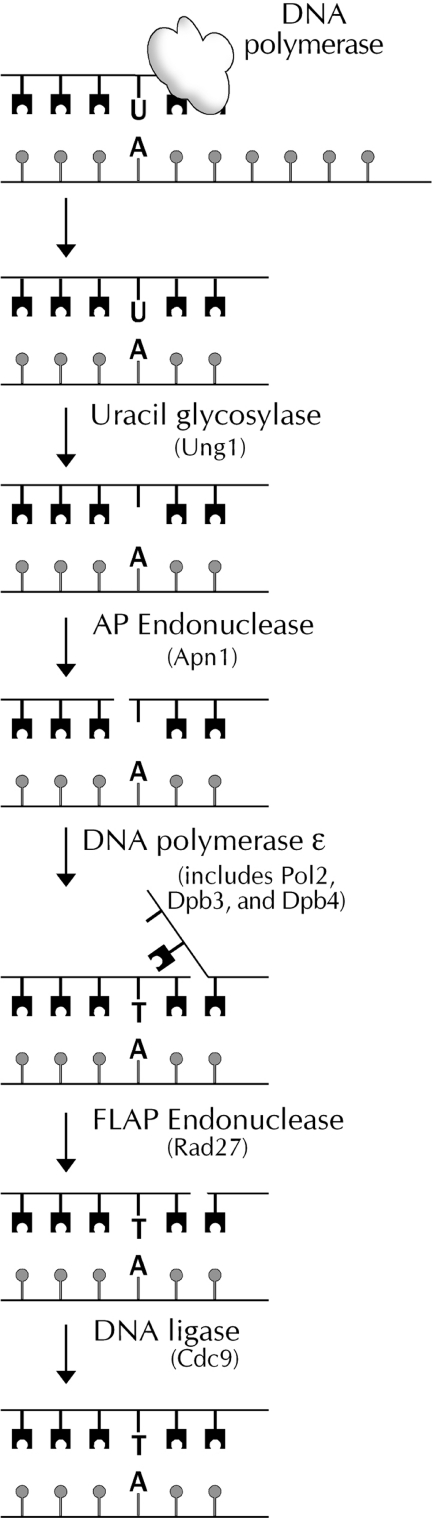
The repair of deoxyuridine from DNA has been extensively studied both in vitro and in vivo and serves as a model for base excision repair (BER) (12,13). Key features of the repair pathway in S.cerevisiae are shown in Figure 1. To clarify the role of DNA repair in mediating the toxicity of thymidine deprivation, the response to aminopterin, an antifolate, was determined in a series of mutants deficient in uracil BER.
Figure 1.
Schematic view of uracil base excision events occurring after incorporation of dUTP in place of dTTP into DNA. The S.cerevisiae protein thought to be most active at each step is noted in parentheses. See text for further details.
MATERIALS AND METHODS
Strains
S.cerevisiae strains from two backgrounds were used in these studies. Parental and mutant strains in the BY4741 background were obtained from Open Biosystems (Huntsville, AL). Mutants in this strain have been described elsewhere (14) and contain deletions extending from the start codon through the stop codon for the gene of interest. The deleted endogenous gene is replaced with a neomycin resistance gene.
Additional studies were carried out in strains derived from SSL204 his3 ade2 leu2 ura3 trp1 (15) as indicated. Mutations in various repair genes were introduced into this strain using lithium transformation of linear DNA (16). Deletion alleles replaced by the neomycin resistance gene from the BY4741 background were transferred into the SSL204 background by transforming linear DNA obtained by PCR using primers 1 kb upstream and 1 kb downstream of the transferred locus. When required, the locations of the PCR primers were adjusted to amplify sequences outside genes adjacent to the transferred locus. Deletion alleles marked with HIS3 or TRP1 were created by using the method described by Burke et al. (16,17). Linear DNA for transformation was created by PCR utilizing 60 base primers containing 40 bases complementary to the immediate 5′ or 3′ coding sequence of the gene to be deleted. The 3′ 20 bases of the primers were complementary to sequences flanking either the HIS3 or TRP1 gene in plasmids pRS313 or pRS424 (18).
Aminopterin treatment
Cultures were grown overnight in minimal medium supplemented with 0.2% norite treated casamino acids and 20 µg/ml of tryptophan, adenine and uracil, pH 6.0. Cultures were diluted 1:100 into fresh medium and outgrown 4 h. Cell density was then determined, and 105 cells/ml were placed into fresh medium described above but also containing 6 mg/ml sulfanilamide and 0.2 mg/ml aminopterin (Sigma Aldrich, St Louis, MO). Sulfanilamide was added to medium prior to autoclaving (19). Aliquots from the culture were removed at indicated times, diluted in water and plated to solid YPD (yeast extract, peptone and dextrose) medium. Cultures without drug were established and plated in parallel to ensure viability of the cells.
Methylene blue
Cells from treated and untreated cultures were stained with methylene blue as described by Sami et al. (20).
Flow cytometry
Cultures were grown as described above, except the drug treatment was performed at a cell density of 106 cells/ml. At various time points, 107 cells were removed, washed in water and resuspended in 70% ethanol. Cells were prepared similarly to the procedure described by Paulovich and Hartwell (21). Briefly, cells were collected, resuspended in water, collected again, and resuspended in 70% ethanol. Cells were then incubated at 4°C on a rotating table for 3–24 h before being collected and resuspended in 50 mM sodium citrate buffer (pH 7.5) and treated with RNase A and proteinase K. The cells were incubated with propidium iodide overnight and then sonicated. Cells were analyzed in a Becton Dickson FACscan.
QPCR assay
Total DNA was extracted from aminopterin-treated samples using a MasterPure™ Yeast DNA Purification Kit (Epicentre, Madison, WI) according to the manufacturer's instructions. DNA concentration was determined with the Quant-iT™ PicoGreen® dsDNA Assay Kit (Molecular Probes, Eugene, OR), adjusted to 3 ng/µl, and confirmed with PicoGreen on a SPECTRAFluor Plus microplate reader (Tecan, Research Triangle Park, NC) at an excitation wavelength of 485 nm and emission of 535 nm. DNA damage was analyzed by a quantitative PCR method adapted from Santos et al. (22), followed by PicoGreen staining. The oligo pair designed to amplify a 9.3 kb region of chromosome XIII including genes PFK2 and HFA1 were 5′-CAA AGA ACC GTC ACC ACC ACA AA-3′ and 5′-CGC TAA AAT CCC GTG TAT CCC TTG-3′. Quantitative PCR was performed with the GeneAmp® XL PCR Kit (Applied Biosystems, Foster City, CA) on an MJ Research PTC-200 DNA Engine (Bio-Rad Laboratories, Inc., Waltham, MA). The reaction mixture contained 12 ng of DNA, 1× XL Buffer II, 100 ng/µl BSA, 200 µM of dNTPs, 10 pmol of each primer and 0.8 mM of Mg2+. Following a hot start, 0.2 U of rTth polymerase diluted in water were added. Cycling conditions were as follows: hot start at 75°C for 2 min, initial denaturation at 94°C for 1 min, followed by 22 cycles of 15 s at 94°C and 12 min at 65°C, and a final 10 min extension at 72°C. PCR products were initially visualized by ethidium bromide gel electrophoresis and then analyzed by PicoGreen quantitative analysis.
RESULTS
Changes in dUTP to TTP ratio, uracil incorporation into DNA and subsequent repair events to excise and replace uracil may all potentially contribute to the toxicity of thymidine depletion. This study explores the connection between uracil-DNA BER (shown in Figure 1) and thymidine deprivation toxicity by examining the sensitivity of mutants in uracil-DNA BER to aminopterin. Aminopterin was used to inhibit dihydrofolate reductase and sulfanilamide to inhibit de novo synthesis of reduced folate. Together, these agents induce thymidine deprivation by removing the source of the one carbon group transferred to dUMP by thymidylate synthase to form dTMP (19). Isogenic strains of S.cerevisiae differing at nonessential loci involved in uracil-DNA repair were treated with aminopterin as described in Materials and Methods. The survival of cultures following a 24 h exposure to aminopterin and sulfanilamide is shown in Table 1. In wild-type BY4741 cells under the conditions used here, a 24 h exposure led to 23% survival. Uracil glycosylase (ung1) mutants and rad27 mutants showed a slightly altered sensitivity to aminopterin with survival of 36 and 2.3%, respectively. The rad27 mutants formed adherent clusters resistant to sonication in response to aminopterin, thereby probably giving an underestimate of survival by the clonogenic survival assay. DNA ligase null mutants are not viable and were therefore not examined. DNA polymerase ɛ has been implicated in DNA repair synthesis following uracil excision (13). Cells without this polymerase activity are not viable; however, some subunits of the polymerase can be deleted without loss of viability. Mutants lacking a subunit of polymerase ɛ are viable but have increased mutation rates (23,24). These subunit mutants, dpb3 and dpb4, showed a modest increase in sensitivity to aminopterin relative to wild-type cells.
Table 1.
Survival after 24 h in Aminopterin plus Sulfanilamide medium
| Relevant genotype | % survival, 24 h | SD | t-Test |
|---|---|---|---|
| (A) BY4741 backgrounda | |||
| Wild-type | 22.8 | 8.7 | |
| ung1 | 36.0 | 6.6 | 0.05 |
| apn1 (6 h) | 1.1 | 0.06 | 0.001b |
| apn1 (24 h) | 0.08 | 0.008 | 8.8 × 10−8 |
| apn2 | 39.3 | 18 | 0.25 |
| pol32 | 33.3 | 21 | 0.49 |
| rad27 | 2.3 | 0.6 | 2.6 × 10−7 |
| dpb3 | 13.3 | 0.6 | 9.0 × 10−4 |
| dpb4 | 9.3 | 4.9 | 0.014 |
| (B) SSL204 backgroundc | |||
| Wild-type | 10.8 | 4.4 | |
| ung1 | 11.2 | 5.4 | 0.9 |
| apn1 | 0.03 | 0.01 | <0.05 |
| rad27 | 11.0 | 5.8 | 0.95 |
| (C) apn1 combination mutants in SSL204 backgroundd | |||
| apn1 ung1 | 12.7 | 1.5 | 0.005 |
| apn1 ntg1 ntg2 | 0.04 | 0.01 | 0.54 |
| apn1 rev3 (12 h)e | 0.10 | 0.01 | 0.82 |
| apn1 apn2 | 0.09 | 0.04 | 0.33 |
aVarious mutants in the uracil-BER pathway in the BY4741 background were treated with aminopterin and sulfanilamide as described in Materials and Methods. Following a 24 h exposure, portions of the culture were collected and plated to rich YPD medium. Colonies were counted after 4 days. Survival is reported as the number of colony forming units per milliliter at 24 h/colony forming units per milliliter at time zero. apn1 mutants were assessed at earlier time points given the lack of sufficient survivors at 24 h.
bCompared with wild-type at 6 h.
cAminopterin sensitivity was determined as described above for mutants generated in the SSL204.
dAminopterin sensitivity of apn1 mutant strains bearing mutations in additional DNA repair genes.
eCompared with apn1 at 12 h.
In contrast to the modest changes in sensitivity seen for the above mutants, we found a profound sensitivity to aminopterin in apn1 mutants. At 6 h of aminopterin treatment, wild-type and other mutants examined showed a plateau in the concentration of viable cells in the culture. In contrast, apn1 mutant cultures demonstrated a survival of 1%. Thymidine deprivation for 24 h led to a >250-fold difference in survival between wild-type and apn1 mutants. This is the largest difference in survival reported for any single mutation in S.cerevisae for thymidine deprivation to the best of our knowledge.
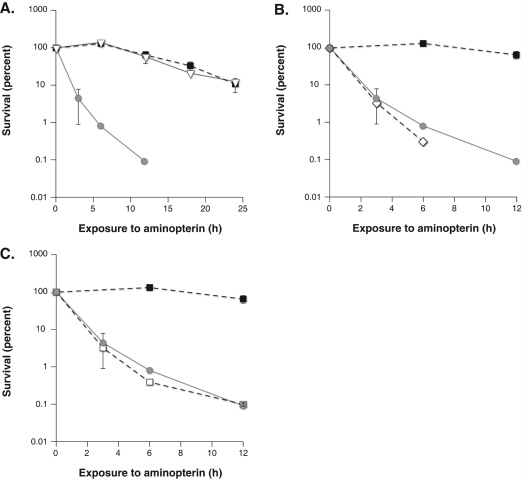
In order to confirm these findings in an independent strain of S.cerevisiae and address the possibility of strain background contributing to these findings, the apn1, ung1 and rad27 genes in parental strain SSL204 (15) were disrupted [Figure 2 and Table 1, (B)]. Using the SSL204 derivatives, extreme sensitivity in the apn1 mutant was again observed, highlighting the potential importance of Apn1 in mediating the response to thymidine deprivation. The SSL204:rad27 mutant did not aggregate upon exposure to aminopterin and had a survival rate comparable with the SSL204 parental strain by the clonogenic survival assay. These findings suggest that the role of Rad27 in mediating the toxicity of thymidine deprivation may be limited. The extreme sensitivity of apn1 deletion seen in both strains suggests that blocking the BER pathway specifically at the abasic incision step is extremely toxic to thymidine deprived cells.
Figure 2.
(A) Survival of wild-type, apn1 ung1 and apn1 strains in the SSL204 background. Cultures of the indicated genotype were treated with aminopterin and sulfanilamide. At various times portions of the cultures were removed and plated to rich YPD medium. Colonies were counted after 4 days. Survival is determined as the number of colony forming units at a given time relative to the initial concentration of colony forming units per milliliter. The sensitivity of apn1 mutants is relieved in apn1 ung1 double mutants. (B) The sensitivity of apn1 mutants is not relieved in apn1 ntg1 ntg2 triple mutants. Wild-type, apn1 and apn1 ntg1 ntg2 mutants were treated with aminopterin and sulfanilamide as described in Materials and Methods. Survival was determined by plating portions of the culture to rich medium at various times of drug exposure. (C) The sensitivity of apn1 mutants is not relieved in apn1 rev3 double mutants. Survival was determined as described previously. SSL204 parental strain is closed square, apn1 is gray circle, apn1 ung1 mutant strain is open triangle, apn1 ntg1 ntg2 is open diamond and apn1 rev3 is open squares.
The UNG1 gene in an apn1 mutant was disrupted to further characterize the sensitivity of apn1 mutants. Deletion of uracil glycosylase should prevent the formation of abasic sites and prevent the extreme toxicity seen in apn1 mutants. The double apn1 ung1 mutant showed aminopterin sensitivity comparable with wild-type cells, demonstrating significant protection of apn1 mutant cells by the additional loss of uracil glycosylase activity [Figure 2 and Table 1, (C)]. The protection afforded by deletion of the uracil glycosylase gene confirms that attempted repair of uracil is responsible for the severe toxicity of aminopterin in apn1 mutants.
One potential explanation for the aminopterin sensitivity seen in apn1 mutants is that abasic sites formed during uracil-BER become substrates for less efficient or outright toxic alternate repair pathways. Attempting DNA repair during thymidine depletion may result in entry into a repair pathway that cannot be completed. Since BER of uracil involves strand cleavage, stalled or incomplete repair events may be more toxic than the initial repair substrate, uracil-DNA. Abasic sites can be repaired by alternative pathways (13), particularly in the absence of Apn1. Two alternative pathways involved in abasic DNA metabolism were examined. Ntg1 and Ntg2 are S.cerevisiae homologs of E.coli endonuclease III and, like endonuclease III, are BER enzymes with broad specificity. Ntg1 and Ntg2 have an associated AP lyase activity which, unlike AP endonuclease, cleaves 3′ to the AP site in a beta elimination reaction, leaving a 3′ α–β unsaturated aldehydic (3′dRP) end. Strand cleavage by this mechanism thus can create toxic 3′ blocked ends unsuitable for priming DNA repair synthesis. Hanna et al. (25) showed that S.cerevisiae mutants lacking AP endonuclease activity are very sensitive to DNA methylation damage normally repaired by BER. They further showed that this sensitivity can be relieved by deletion of NTG1 and NTG2 and concluded that in the absence of functional AP endonuclease activity, abasic sites are acted upon by the lyase activity of Ntg1 and Ntg2 to create toxic repair intermediates. Therefore, inactivating NTG1 and NTG2 avoided these intermediates and allowed for increased survival during methylation damage (25). To determine if similar pathways were operating during thymidine starvation, a triple mutant strain with deletions in APN1, NTG1 and NTG2 was created and found to remain very sensitive to aminopterin (Figure 2). The sensitivity of the triple mutant suggests that the extreme sensitivity of apn1 mutants is not due to shuttling abasic repair intermediates into a pathway which creates more toxic intermediates via Ntg1 or Ntg2 abasic lyase activity.
Abasic sites can also be bypassed by translesional synthesis which may be mutagenic and could result in accumulation of a large number of potentially toxic mutations. To determine if the sensitivity of apn1 mutants was due to translesional synthesis, the aminopterin sensitivity of apn1 rev3 double mutants was examined. Rev3 is a component of polymerase ζ, known to be active in error prone translesional synthesis. The double mutants were essentially as sensitive to aminopterin as apn1 single mutants (Figure 2). This suggests that the extreme sensitivity of apn1 mutants is not due to shuttling of abasic site lesions into a translesional bypass pathway. Taken together, these observations suggest that unrepaired abasic sites or the block imposed by apn1 deletion are extremely toxic in thymidine starved cells.
S.cerevisiae contains an additional AP endonuclease, Apn2, identified by its homology to other AP endonucleases (26). apn2 deletion mutants have a sensitivity to aminopterin similar to wild-type cells [Table 1, (A)]. The sensitivity of apn1 and apn1 apn2 mutants in the SSL204 background was determined. After 24 h of aminopterin, the apn1 mutant showed 0.03% (SD 0.000 2) survival and the apn1 apn2 double mutant showed 0.09% survival (SD 0.04, not significantly different P > 0.05). The parental SSL204 survival is 10.8%. The double apn1 apn2 mutant has a sensitivity similar to the apn1 mutant, suggesting that Apn2 activity is not responsible for creating toxicity during aminopterin treatment in apn1 mutant cells.
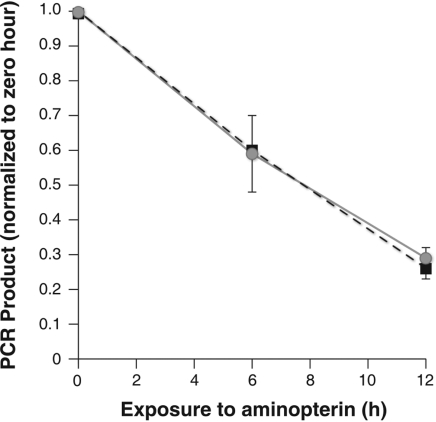
The genetic analyses shown above failed to provide evidence that alternative abasic repair contributes to the toxicity seen in apn1 mutants. To assess the possibility that DNA damage not mediated by Apn2, Ntg1, Ntg2 or Rev3 was responsible for the sensitivity of apn1 mutants, DNA from aminopterin treated cells was examined. Since several types of DNA damage may be initiated by thymidine deprivation, including single and double strand breaks, stalled replication and recombination intermediates and potentially DNA–protein cross links (27–29), DNA damage was assessed with a non-discriminatory, broad specificity PCR assay described by van Houten et al. (30) and shown to be effective in determining the extent of DNA damage from divergent sources including cisplatin, oxidative injury and UV light. In this assay, DNA purified from cells prior to and following aminopterin treatment is quantified and used as template to produce a 9.3 kb fragment from chromosome XIII (22). Conditions for the reaction are standardized such that the amount of PCR product DNA is proportional to the amount of intact template DNA present in the reaction. Double strand and single strand breaks decrease the efficiency of the reaction. Other DNA lesions, such as abasic sites, also interfere with the progress of DNA polymerase along the template and will decrease the amount of product. During aminopterin exposure, the amount of viable template decreases with time equally in wild-type and apn1 mutant cells (Figure 3). The nature of the obstructing lesion in aminopterin treated cells cannot be determined by this assay; however, the PCR findings suggest that the significant sensitivity of apn1 mutants is not due to preferential DNA degradation or damage in apn1 mutants.
Figure 3.
Wild-type and apn1 mutants were exposed to aminopterin and sulfanilamide as described previously. Portions of the culture were collected prior to drug exposure, at 6 h and at 12 h, and DNA was purified as described in Materials and Methods. PCR was performed using the conditions and primers characterized by van Houten (30). PCR product was quantified using the PicoGreen assay. The amount of product PCR at a given time-point was normalized to the amount of product derived from input DNA from the pre-drug treatment DNA. Parental SSL204 is closed squares and SSL204 apn1 mutants are gray circles.
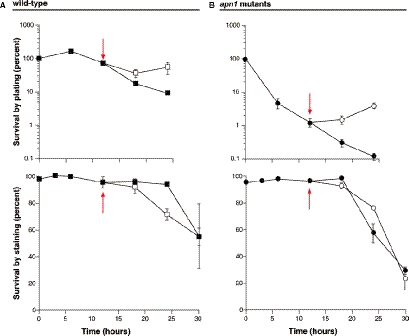
The above studies show apn1 mutants are uniquely sensitive to aminopterin among uracil-BER mutants. DNA damage and/or repair probably plays a role in the sensitivity of apn1 mutants since their sensitivity is absent in the ung1 apn1 double mutant. Since apn1 mutants are much more sensitive to aminopterin than wild-type cells, yet show comparable DNA damage during exposure, the effects of aminopterin exposure on viability of wild-type and apn1 mutants after removal of the drug and during recovery were examined. The colony formation assays used above cannot determine precisely when the toxicity of thymidine deprivation occurs; therefore, methylene blue viability staining was used to characterize the timing of aminopterin toxicity more closely. Cultures of both wild-type and apn1 mutants were treated with aminopterin as in previous experiments with the exception of increasing the cell concentration to 106 cells/ml. The higher cell density allowed collection of adequate numbers of cells for additional flow cytometry studies described below. At various times, portions of the cultures were removed and stained with methylene blue. Although both living and dead cells take up methylene blue, only living cells are able to convert the dye to a colorless compound, leaving dead cells blue (20). At the same time points, portions of the cultures were also plated to rich medium to determine viability via clonogenic survival as in prior experiments. At 12 h, cells from a portion of the drug treated culture were collected, washed and resuspended in an equal volume of fresh medium without aminopterin and sulfanilamide. The results are shown in Figure 4. The effects of aminopterin on colony formation during exposure were similar to the effects described above (Figure 4A and B). In contrast to the viability determined by plating, viability determined by methylene blue staining remained high for both strains until after 18 h after of drug exposure. At times later than 18 h, cultures for both wild-type and apn1 mutants began to show loss of viability by methylene blue staining. This observation suggests the toxicity of thymidine starvation may not be as immediate as the colony formation data suggest. In other words, 12 h of aminopterin exposure leads to decreased colony formation ability but does not result in immediate cytotoxicity. The cultures that had been switched to drug-free medium showed a continued loss of viability by methylene blue staining even after removal from aminopterin. Viability by plating remained essentially stable for both wild-type and apn1 mutants, 6 h after removal of aminopterin. The differences between clonogenic survival and methylene blue viability and the observation that toxicity by staining continues despite a switch to drug-free medium suggest that at least a portion of toxicity resulting from aminopterin treatment occurs after and not during drug exposure.
Figure 4.
Wild-type and apn1 mutants were exposed to aminopterin and sulfanilamide as described previously, except in this experiment the cell concentration of exposed cultures was increased to 106 cells/ml. In addition, at 12 h, a portion of the culture was collected, washed and resuspended in fresh medium without drug. A portion of the culture was also maintained in drug-containing medium. Survival was determined as described previously and is shown in (A and B). To determine the viability of cells immediately after treatment, a portion of the culture was stained with methylene blue and 100 cells were counted. Cells with blue cytoplasm were scored as dead, while those with clear cytoplasm were scored as alive. Stationary phase cells were used as controls for living cells and ethanol treated cells were used as controls for dead cells. Methylene blue staining shows a slight decrease in viability for apn1 mutants following 24 h in aminopterin and sulfanilamide but also suggests much of the toxicity of drug treatment occurs after drug exposure. Closed symbols represent continuously treated cultures; open symbols represent cultures treated with drug before being switched to drug-free medium at 12 h. Wild-type cells are squares; circles are apn1 mutants.
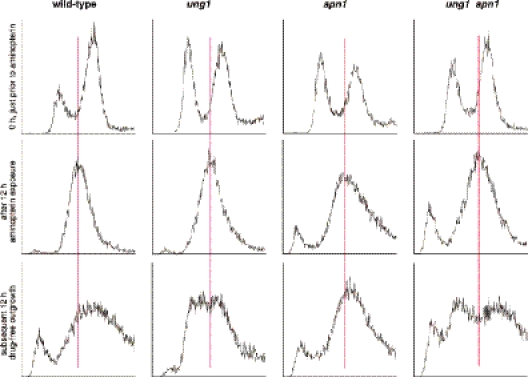
To further characterize changes in DNA during and after aminopterin exposure, treated and recovering cells were examined by flow cytometry. Thymidine depletion, similar to hydroxyurea induced dNTP depletion, is known to create an S-phase arrest (31). Mutants unable to execute a fully functional arrest are sensitive to dNTP depletion resulting from hydroxyurea exposure (32). If indeed a significant portion of the toxicity of thymidine deprivation occurs after and not during drug exposure, cell cycle arrest and recovery kinetics in wild-type and apn1 mutants may be informative. A portion of the cultures examined in the above methylene blue experiment (Figure 4) were collected and analyzed by flow cytometry to determine DNA content changes in response to aminopterin treatment and drug-free regrowth. The results are shown in Figure 5. The response to aminopterin in ung1 mutants and apn1 ung1 double mutants by flow cytometry was also determined. The results in Figure 5 show wild-type, apn1, ung1 and apn1 ung1 double mutant cells all arrested in S phase during aminopterin treatment, consistent with its activity as a DNA replication blocking agent. Upon removal of drug, wild-type cells were able to re-initiate DNA synthesis and re-enter the cell cycle. apn1 mutants, though viable by methylene blue assay, were not able to re-enter the cell cycle and remained fixed in S phase. This observation suggests the severe toxicity of aminopterin in apn1 mutants may result from unrepaired abasic sites blocking re-initiation of replication. Cells lacking uracil glycosylase (ung1) were able to re-initiate DNA synthesis when placed in drug-free medium following aminopterin treatment. Double mutants lacking both uracil glycosylase and Apn1 were able to re-enter the cell cycle with kinetics similar to ung1 mutants. As shown in Figure 2, the double mutant also showed significantly less aminopterin toxicity than apn1 mutants. The extreme sensitivity noted for apn1 mutants therefore appears to result from blocked recovery following aminopterin exposure and not from events or damage during the drug treatment itself.
Figure 5.
Cells from the experiment described in Figure 4 were collected, fixed in ethanol and stained with propidium iodide. DNA content was determined using flow cytometry. While wild-type cells were able to resume DNA synthesis by 12 h after drug removal, apn1 mutants remained firmly fixed in S-phase. The vertical line serves as a reference for the cell cycle position seen in the culture following 12 h of aminopterin treatment.
DISCUSSION
The experiments described here examine the role of uracil BER in mediating the toxicity of thymidine deprivation. Mutants deficient in various aspects of uracil BER were determined to have altered sensitivity to thymidine depletion. While most mutants in the uracil-BER pathway have subtle effects on the toxicity of aminopterin mediated thymidine depletion, apn1 mutants show a profound sensitivity. A modest increase in sensitivity in apn1 mutants to the antifolate trimetrexate has also been described by Simon et al. (33). Aminopterin toxicity in wild-type cells and to a larger extent in apn1 mutants appears to occur after drug exposure and is potentially owing to blocked re-initiation of replication.
Thymidine deprivation causes a relative rise in dUTP that has been suggested to overwhelm dUTPase (34,35). A role for dUTP in thymidineless death is supported by the findings that S.cerevisiae with decreased dUTPase activity are more sensitive to antifolate treatment (4) and human cancer cell lines expressing elevated amounts of dUTPase are resistant to thymidine deprivation (36,37). Gadsden et al. (10) found the toxicity of thymidine depletion in dut1 mutants of S.cerevisiae was exacerbated in the dut1 ung1 double mutant, a background favoring stable accumulation of uracil in DNA. In addition, while the dut1 mutants died in S phase during thymidine depletion (by cell morphology), the dut1 ung1 mutants died throughout the cell cycle (unbudded, large and small budded cells), leading the authors to conclude that excessive dUTP accumulation led to a generalized failure in macromolecular synthesis. It should be noted that only dut1 ung1 double mutants showed this phenotype, implying that global transcriptional failure may result only under conditions of low TTP and very high dUTP afforded by the absence of dUTPase. The extreme sensitivity of apn1 mutants demonstrated here suggests a mechanism involving repair and/or cell cycle arrest rather than alterations in global transcription rates.
Rampant DNA degradation resulting from attempted repair of uracil laden DNA has also been suggested as a mechanism of thymidineless death. However, manipulations of uracil glycosylase alone have not resulted in major effects on the toxicity of thymidine deprivation either in S.cerevisiae or human cancer cells (4,8,36,37). Excess DNA damage does not appear to be responsible for the significant sensitivity of apn1 mutants. Measures of DNA integrity, including the PCR assay and flow cytometry, failed to show increased DNA damage in apn1 mutants. In addition, deletion of genes for repair enzymes with AP lyase activity, NTG1 and NTG2, did not relieve the sensitivity of apn1 deletion. Although inactivating these genes may prevent formation of even more toxic repair intermediates and has been shown to provide protection of apn1 apn2 double mutants during methylation damage (25), no survival advantage was seen in apn1 ntg1 ntg2 triple mutants. Altogether, the data do not support excess DNA damage as an explanation for the extreme sensitivity of apn1 mutants.
A third possible mechanism for thymidineless death suggests enzymes involved in uracil-BER interfere with the delicate process of S-phase arrest, replication fork stabilization and replication re-initiation when thymidine is again available. Nucleotide pool imbalance leads to replication fork stalling and cell cycle arrest by inducing S-phase checkpoints. This process has been studied extensively in yeast using hydroxyurea, which depletes all dNTPs by inhibiting ribonucleotide reductase. Mutants defective in S-phase arrest and replication fork stabilization are very sensitive to hydroxyurea (32), highlighting the importance of these functions. Thymidine depletion also causes cell cycle arrest in S phase (4) and Rad53 activation (31). Mutants deficient in appropriate S-phase arrest, e.g. mec1 and mec2 mutants, are sensitive to both hydroxyurea and methotrexate (33). apn1 mutant cells, in contrast to wild-type cells, remained fixed in S-phase with virtually no cell cycle progression 12 h after aminopterin removal. apn1 mutants remained as viable as wild-type cells after 12 h in aminopterin as assessed by methylene blue viability staining. The lack of S-phase recovery seen in apn1 mutants may be responsible for the toxicity in these cells.
Unlike hydroxyurea, aminopterin treatment under the conditions used in the experiments described here depletes only thymidine and as mentioned above, dUTP may allow cells to transiently overcome the thymidine deficiency (4). The use of dUTP as a thymidine analog may allow more replication to occur than during hydroxyurea treatment, but at the expense of increased dUTP incorporation. The unique toxicity of thymidine deprivation may result from interference with S-phase checkpoints and stability of stalled replication forks by uracil-BER proteins. Uracil glycosylase itself may interfere with appropriate arrest or recovery by binding to either uracil-DNA or abasic sites. Uracil glycosylase has been demonstrated to localize to replication forks (38). In addition, Parikh et al. (39) have shown that uracil glycosylase has a higher affinity for abasic sites than for uracil-DNA. It has been suggested that BER enzymes interact with each other to shield DNA repair intermediates from entering into more toxic manipulations (40). AP endonuclease may serve to accelerate the exit of uracil glycosylase from its product AP site and thereby expedite the repair pathway. In vitro data have shown that AP endonuclease facilitates the dissociation of uracil glycosylase from its substrate and that the presence of AP endonuclease stimulates the base excision activity of uracil glycosylase (39). If Apn1 is absent, uracil glycosylase may remain resident at the uracil-DNA site, inhibiting further processing of the lesion either by repair or replication restart. Human thymine-DNA glycosylase has also been shown to bind tightly to abasic DNA produced by glycosylase action (41). Our observation that an ung1 deletion rescues the extreme sensitivity of apn1 mutants is consistent with this possibility. The toxicity of dUTP incorporation may result more from uracil-BER intermediates blocking recovery from S-phase arrest than from strand cleavage and DNA degradation.
Disruption of BER in mammalian cells also influences the toxicity of thymidine depletion. Li et al. (42) used wild-type and DNA polymerase β knockout murine embryo fibroblasts to show mutants deficient in DNA repair synthesis are resistant to thymidine depletion. As shown in yeast in this study, arrest kinetics during drug exposure were similar for both repair proficient and deficient MEF cells. The repair synthesis mutants were able to re-initiate replication more quickly than wild-type cells. These findings again illustrate that repair activities occurring after removal of thymidine synthesis blockade determine the toxicity of thymidine deprivation. The effect on survival from alterations in BER appear to be dependent on which BER step is altered.
The profound toxicity engendered by loss of Apn1 seen in these experiments suggests the possibility that manipulation of Apn1 in mammalian cells may improve the effectiveness of chemotherapy agents that act via thymidine depletion. Berry et al. (43) used methoxylamine as an inhibitor of BER to alter sensitivity of human and mouse cells to iododeoxyuridine. Using a growth inhibition assay, the group was able to show that methoxylamine further accentuates the growth inhibitory effects of IUdR. Data from the present study suggest arrest and re-initiation of replication contribute significantly to the toxicity of thymidine deprivation, and therefore proliferation assays may provide a more accurate assessment of efficacy compared with growth inhibition assays. Methoxylamine inhibits completion of BER by binding to the DNA substrate (44) and blocking strand incision. An alternative strategy would be to inhibit Apn1 directly, allowing uracil glycosylase to bind to uracil laden DNA and inhibit re-entry into the cell cycle. The possibility that AP endonuclease inhibition at the enzyme level potentiates the cytotoxicity of thymidine depletion in human cancer cells is currently under investigation.
The data presented in this paper suggest that the toxicity of thymidine deprivation occurs in large part after the thymidine depleting agent has been removed and that re-initiation of replication is a particularly sensitive event during thymidine depletion. Interruption of uracil-BER at the abasic endonuclease step produced extreme cytotoxicity. These findings provide a rationale for understanding thymidineless death in the context of interference between uracil-BER and replication arrest and re-initiation. The presence of uracil and uracil glycosylase in the vicinity of stalled replication forks adds to the complexity of replication fork stalling and re-initiation, which has been well described for dNTP depletion via hydroxyurea. The findings also provide a rationale for manipulating abasic endonuclease and S-phase arrest functions during recovery from thymidine depletion as a means to potentiate the cytotoxicity of chemotherapy agents working via thymidine depletion.
Acknowledgments
We appreciate the assistance of Mr Justin Fishbaugh at the University of Iowa Carver College of Medicine Flow Cytometry Core Facility. The authors acknowledge and deeply appreciate the outstanding technical assistance of Ms Kellie Bodeker Goranson with graphics and editing and Ms Suwimol Jetawattana for construction of the apn1 rev3 mutant. We would also like to thank Dr Douglas Spitz for his energetic and wise guidance and Dr Eric Radany for thoughtful discussions on thymidineless death. This work was supported in part by a grant from American Cancer Society through the Holden Comprehensive Cancer Center, University of Iowa. Funding to pay the Open Access publication charges for this article was provided by the authors.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barner H.D., Cohen S.S. The induction of thymine synthesis by T2 infection of a thymine requiring mutant of Escherichia coli. J. Bacteriol. 1954;68:80–88. doi: 10.1128/jb.68.1.80-88.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad S.I., Kirk S.H., Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 3.Ingraham H.A., Dickey L., Goulian M. DNA fragmentation and cytotoxicity from increased cellular deoxyuridylate. Biochemistry. 1986;25:3225–3230. doi: 10.1021/bi00359a022. [DOI] [PubMed] [Google Scholar]
- 4.Tinkelenberg B.A., Hansbury M.J., Ladner R.D. dUTPase and uracil-DNA glycosylase are central modulators of antifolate toxicity in Saccharomyces cerevisiae. Cancer Res. 2002;62:4909–4915. [PubMed] [Google Scholar]
- 5.Richardson C.C., Schildkraut C.L., Kornberg A. Studies on the Replication of DNA by DNA Polymerases. In: Frisch L., editor. Synthesis and Structure of Macromolecules, 1st edn. Vol. XXVIII. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1963. p. 629. Cold Spring Harbor Symposia on Quantitative Biology. [Google Scholar]
- 6.Lawrence T.S., Maybaum J. Fluoropyrimidines as radiation sensitizers. Semin. Radiat. Oncol. 1993;3:20–28. doi: 10.1053/SRAO00300020. [DOI] [PubMed] [Google Scholar]
- 7.Dusenbury C.E., Davis M.A., Lawrence T.S., Maybaum J. Induction of megabase DNA fragments by 5-fluorodeoxyuridine in human colorectal tumor (HT29) cells. Mol. Pharmacol. 1991;39:285–289. [PubMed] [Google Scholar]
- 8.Burgers P.M., Klein M.B. Selection by genetic transformation of a Saccharomyces cerevisiae mutant defective for the nuclear uracil-DNA-glycosylase. J. Bacteriol. 1986;166:905–913. doi: 10.1128/jb.166.3.905-913.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.el-Hajj H.H., Zhang H., Weiss B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 1988;170:1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadsden M.H., McIntosh E.M., Game J.C., Wilson P.J., Haynes R.H. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993;12:4425–4431. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.el-Hajj H.H., Wang L., Weiss B. Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J. Bacteriol. 1992;174:4450–4456. doi: 10.1128/jb.174.13.4450-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parikh S.S., Putnam C.D., Tainer J.A. Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res. 2000;460:183–199. doi: 10.1016/s0921-8777(00)00026-4. [DOI] [PubMed] [Google Scholar]
- 13.Boiteux S., Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Rep. (Amst.) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 15.Ahn B.Y., Livingston D.M. Mitotic gene conversion lengths, coconversion patterns, and the incidence of reciprocal recombination in a Saccharomyces cerevisiae plasmid system. Mol. Cell. Biol. 1986;6:3685–3693. doi: 10.1128/mcb.6.11.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke D., Dawson D., Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, 2000 edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. p. 35. [Google Scholar]
- 17.Burke D., Dawson D., Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, 2000 edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. p. 55. [Google Scholar]
- 18.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickner R.B. Mutants of Saccharomyces cerevisiae that incorporate deoxythymidine-5(-monophosphate into deoxyribonucleic acid in vivo. J. Bacteriol. 1974;117:252–260. doi: 10.1128/jb.117.1.252-260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sami M., Ikeda M., Yabuuchi S. Evaluation of the alkaline methylene-blue staining method for yeast activity determination. J. Ferment Bioeng. 1994;78:212–216. [Google Scholar]
- 21.Paulovich A.G., Hartwell L.H. A checkpoint regulates the rate of progression through S phase in S.cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 22.Santos J.H., Mandavilli B.S., van Houten B. Measuring oxidative mtDNA damage and repair using quantitative PCR. Methods Mol. Biol. 2002;197:159–176. doi: 10.1385/1-59259-284-8:159. [DOI] [PubMed] [Google Scholar]
- 23.Ohya T., Maki S., Kawasaki Y., Sugino A. Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:3846–3852. doi: 10.1093/nar/28.20.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Araki H., Hamatake R.K., Morrison A., Johnson A.L., Johnston L.H., Sugino A. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4867–4872. doi: 10.1093/nar/19.18.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna M., Chow B.L., Morey N.J., Jinks-Robertson S., Doetsch P.W., Xiao W. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Rep. (Amst.) 2004;3:51–59. doi: 10.1016/j.dnarep.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Johnson R.E., Torres-Ramos C.A., Izumi T., Mitra S., Prakash S., Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panadero A., Yin M.B., Voigt W., Rustum Y.M. Contrasting patterns of DNA fragmentation induced by thymidylate synthase inhibitors, ZD1694 and AG-331. Oncol. Res. 1995;7:73–81. [PubMed] [Google Scholar]
- 28.Canman C.E., Tang H.Y., Normolle D.P., Lawrence T.S., Maybaum J. Variations in patterns of DNA damage induced in human colorectal tumor cells by 5-fluorodeoxyuridine: implications for mechanisms of resistance and cytotoxicity. Proc. Natl Acad. Sci. USA. 1992;89:10474–10478. doi: 10.1073/pnas.89.21.10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin M.B., Rustum Y.M. Comparative DNA strand breakage induced by FUra and FdUrd in human ileocecal adenocarcinoma (HCT-8) cells: relevance to cell growth inhibition. Cancer Commun. 1991;3:45–51. [PubMed] [Google Scholar]
- 30.Van Houten B., Cheng S., Chen Y. Measuring gene-specific nucleotide excision repair in human cells using quantitative amplification of long targets from nanogram quantities of DNA. Mutat. Res. 2000;460:81–94. doi: 10.1016/s0921-8777(00)00018-5. [DOI] [PubMed] [Google Scholar]
- 31.Vernis L., Piskur J., Diffley J.F. Reconstitution of an efficient thymidine salvage pathway in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:e120. doi: 10.1093/nar/gng121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lisby M., Barlow J.H., Burgess R.C., Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Simon J.A., Szankasi P., Nguyen D.K., Ludlow C., Dunstan H.M., Roberts C.J., Jensen E.L., Hartwell L.H., Friend S.H. Differential toxicities of anticancer agents among DNA repair and checkpoint mutants of Saccharomyces cerevisiae. Cancer Res. 2000;60:328–333. [PubMed] [Google Scholar]
- 34.Webley S.D., Hardcastle A., Ladner R.D., Jackman A.L., Aherne G.W. Deoxyuridine triphosphatase (dUTPase) expression and sensitivity to the thymidylate synthase (TS) inhibitor ZD9331. Br. J. Cancer. 2000;83:792–799. doi: 10.1054/bjoc.2000.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koehler S.E., Ladner R.D. Small interfering RNA-mediated suppression of dUTPase sensitizes cancer cell lines to thymidylate synthase inhibition. Mol. Pharmacol. 2004;66:620–626. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 36.Welsh S.J., Hobbs S., Aherne G.W. Expression of uracil DNA glycosylase (UDG) does not affect cellular sensitivity to thymidylate synthase (TS) inhibition. Eur. J. Cancer. 2003;39:378–387. doi: 10.1016/s0959-8049(02)00610-x. [DOI] [PubMed] [Google Scholar]
- 37.Canman C.E., Radany E.H., Parsels L.A., Davis M.A., Lawrence T.S., Maybaum J. Induction of resistance to fluorodeoxyuridine cytotoxicity and DNA damage in human tumor cells by expression of Escherichia coli deoxyuridinetriphosphatase. Cancer Res. 1994;54:2296–2298. [PubMed] [Google Scholar]
- 38.Otterlei M., Warbrick E., Nagelhus T.A., Haug T., Slupphaug G., Akbari M., Aas P.A., Steinsbekk K., Bakke O., Krokan H.E. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parikh S.S., Mol C.D., Slupphaug G., Bharati S., Krokan H.E., Tainer J.A. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parikh S.S., Putnam C.D., Tainer J.A. Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res. 2000;460:183–199. doi: 10.1016/s0921-8777(00)00026-4. [DOI] [PubMed] [Google Scholar]
- 41.Hardeland U., Steinacher R., Jiricny J., Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L., Berger S.H., Wyatt M.D. Involvement of base excision repair in response to therapy targeted at thymidylate synthase. Mol. Cancer Ther. 2004;3:747–753. [PubMed] [Google Scholar]
- 43.Berry S.E., Loh T., Yan T., Kinsella T.J. Role of MutSalpha in the recognition of iododeoxyuridine in DNA. Cancer Res. 2003;63:5490–5495. [PubMed] [Google Scholar]
- 44.Liuzzi M., Weinfeld M., Paterson M.C. Selective inhibition by methoxyamine of the apurinic/apyrimidinic endonuclease activity associated with pyrimidine dimer-DNA glycosylases from Micrococcus luteus and bacteriophage T4. Biochemistry. 1987;26:3315–3321. doi: 10.1021/bi00386a011. [DOI] [PubMed] [Google Scholar]