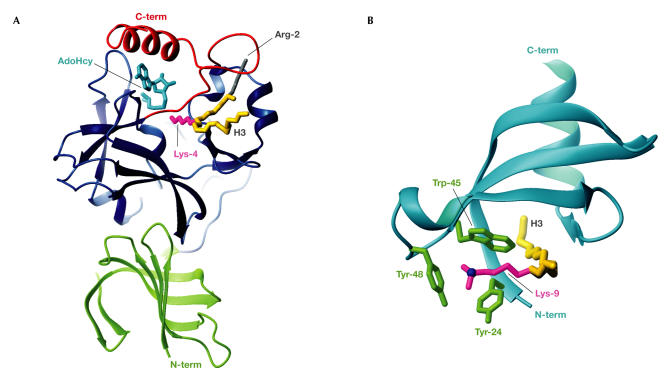
Figure 3.
Methyl-Lys modification and recognition. (A) The SET7/9 domain complexed with AdoHcy (cyan) and an H3 peptide spanning Arg 2 to Lys 9 (backbone in gold; Protein Data Bank (PDB) code ). The H3 methyl-Lys 4 side chain (magenta) projects from the front face, through the lysine-access channel, to reach AdoHcy at the back. The SET domain (blue) contains the conserved SET-N and SET-C domains, each of three or four β-strands, a short helix and several loops, plus the variable SET-I region. The N-terminal SET7/9 domain (green) makes hydrophobic interactions with the SET domain. The SET-C flanking region (red) packs against the SET domain to form the lysine-access channel, the histone-binding groove that selects H3 through interactions with Arg 2 (grey) and hydrophobic interactions with AdoHcy, which collectively explain its importance for SET activity. (B) The HP1β chromodomain (cyan) complexed with an H3 peptide (Gln 5 to Ser 10; backbone in gold) sandwiched between strands β4 and the histone-induced strand β1 (PDB code ). The H3 di-methyl-Lys 9 side chain (magenta) is surrounded along its length by hydrophobic residues that are conserved in many chromodomains. The Lys-Nζ atom (blue) sits centrally in the three-walled aromatic cage that is formed by Tyr 24, Trp 45 and Tyr 48 (green), which can accommodate di- or tri-methyl-Lys.