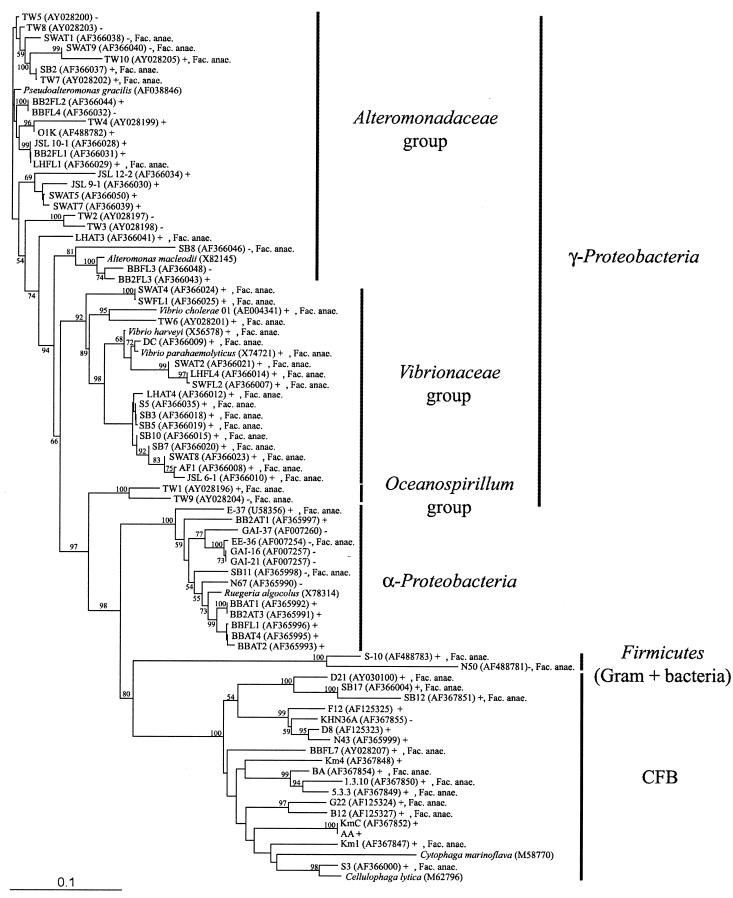
FIG. 1.
Phylogenetic tree showing relationships of the analyzed isolates to representative bacterial 16S rRNA genes obtained from GenBank. The tree was inferred by Clustal W and the neighbor-joining method of on average approximately 400 bp. The number of bootstrap replicates supporting the branching order, from a total of 1,000 replicates, is shown at the segments. Only values of >50% are shown. Symbols and abbreviation: +, NAG uptake, complete or partial inhibition by STZ; −, no NAG uptake, no effect of STZ; Fac. anae., facultative anaerobes. GenBank accession numbers are given after the isolate names. All isolates were isolated from off Scripps Pier except for isolates with the following prefixes: SB, Santa Barbara, California coast (64); JSL, Johnson Sea Link I Research Submersible, off Florida coast; GAI, E, and EE, Georgia coast (22, 23); V. cholerae N16961 (40), V. parahaemolyticus strain BB22 (9), and V. harveyi strain BB120 (8). Isolate AA was identical to MED 11 (AF02554). The scale bar indicates base pair substitutions per nucleotide position.