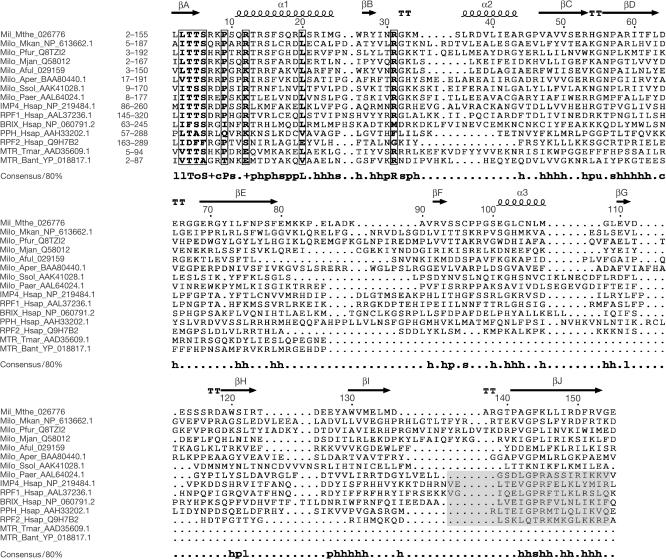
Figure 1.
Sequence alignment of Mil, selected archaeal orthologues, representatives of the five families of eukaryotic Imp4/Brix proteins and the N-terminal regions of predicted bacterial methyltransferases. The eukaryotic families are represented by the human members. The alignment was constructed using the MACAW program (Schuler et al, 1991) and manually refined using the structural elements of Mil as a guide. The structural elements are shown above the alignment, and the consensus including residues conserved in 80% of the aligned sequences is shown underneath the alignment (h stands for hydrophobic residues, l for aliphatic residues, s for small residues, o for alcohol residues, p for polar residues, s for small residues, u for ‘tiny' residues and + for positively charged residues). Boxed residues have a similarity score >0.2, using the BLOSUM62 matrix. The purported σ70-like motif (Wehner & Baserga, 2002) is shadowed in the eukaryotic proteins. For each protein, the range of the aligned residues is indicated. The proteins are designated by their abbreviated name and species name, and GenBank accession number. Aful, Archaeoglobus fulgidus; Aper, Aeropyrum pernix; Bant, Bacillus anthracis; Hsap, Homo sapiens; Milo, Mil orthologue; Mjan, Methanocaldococcus jannaschii; Mkan, Methanopyrus kandleri; Mthe, M. thermoautotrophicus; MTR, methyltransferase; Paer, Pyrobaculum aerophylum; Pfur, Pyrococcus furiosus; PPH, Peter Pan homologue; Ssol, S. solfataricus; Tmar, Thermotoga maritima.