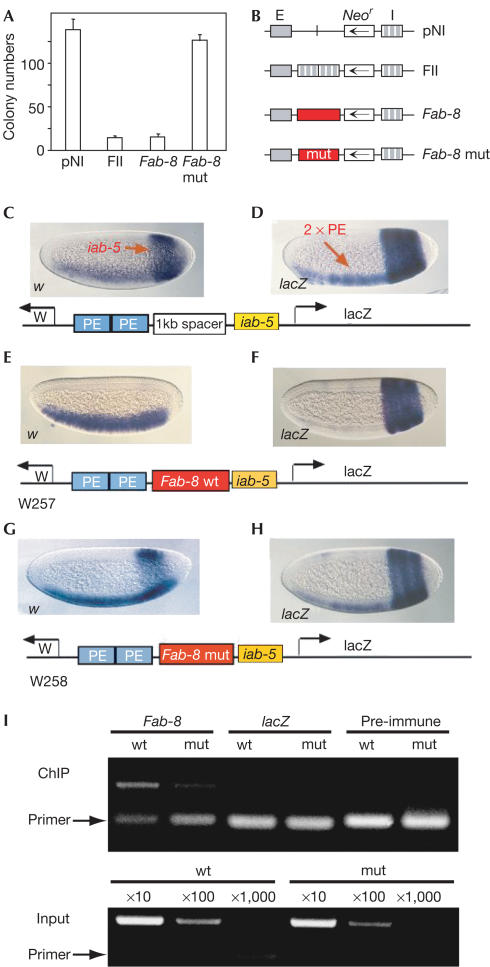
Figure 4.
CTCF-dependent enhancer blocking of Fab-8 in vertebrate cells and in Drosophila. (A) K562 cells were transfected with the indicated DNA constructs (B) and colony number was determined after neomycin selection. The pNI and FII constructs (Chung et al, 1993) were used as controls. Transcription of the neomycin gene (Neo) is driven by the β-globin enhancer (E). In the FII construct, two copies of the β-globin insulator (I) separate the enhancer from Neo. The Fab-8 fragment mediates enhancer blocking, whereas mutation of both CTCF-binding sites (Fab-8 mut) does not. (C–H) Fab-8 enhancer blocking in Drosophila depends on dCTCF-binding sites. Transgenic embryos with the indicated vectors were hybridized with either digoxygenin-labelled white (w) in (C,E,G) or lacZ antisense RNA probes (D,F,H). (C,D) white and lacZ expression is visualized when a 1.6 kb λ DNA fragment was inserted between the 2 × PE and iab-5 enhancers (see text). (E,F) The Fab-8 DNA insert between 2 × PE and iab-5 enhancer was analysed in 13 independent lines. Expression of the white reporter gene is restricted to the ventral mesoderm and of the lacZ gene to the abdomen. With the CTCF sites mutagenized, 22 independent lines were generated: white gene activity in the abdomen (G) and lacZ expression in the ventral mesoderm (H) are seen. (I) Chromatin was prepared from embryos carrying transgenes with either Fab-8 wt or the mut1+2 CTCF-binding sites (mut). Primers that specifically recognize transgenic (not genomic) sequences were used. ChIP using anti-CTCF-C antibody shows an increased occupancy at wild-type versus mutant sequences, in contrast to pre-immune serum and to lacZ sequences. For standardization, input dilutions were subjected to semiquantitative PCR as well.