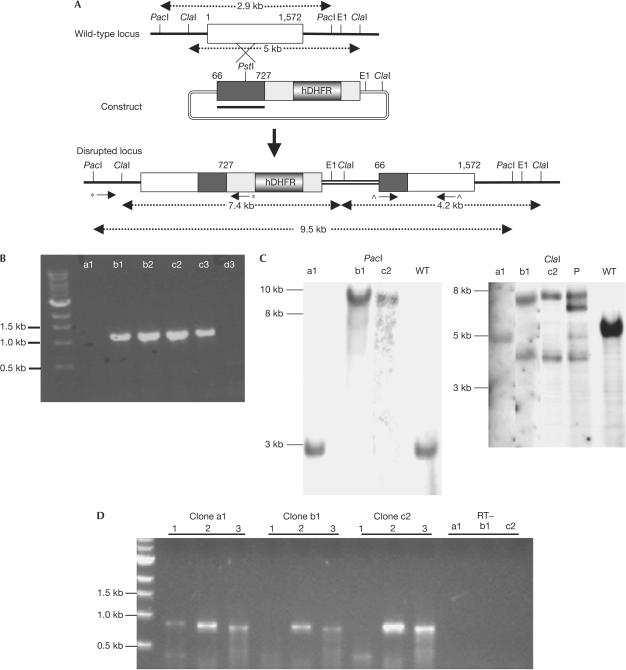
Figure 2.
Strategy for inactivation of the pbmap-2 gene by single cross-over homologous recombination, and genotypic characterization of mutant parasites. (A) Schematic of the wild-type locus, the targeting construct and the disrupted locus. The targeting construct spans positions 66–727 of the 1,572 bp coding sequence (see arrows in Fig 1 of supplementary information online). The disrupted locus contains two nonfunctional copies of the gene. The first copy is truncated at position 727 and lacks essential catalytic residues, whereas the second copy is unlikely to be expressed as it lacks 5′ promoter sequences and the first 65 residues of the coding sequence. (B) Clones recovered by limiting dilution were tested for disruption of the locus by PCR using primers indicated by inverted arrows marked with an asterisk in (A) (primers 1,2; see supplementary Table 3 online for a list of primer sequences). We were able to amplify a fragment of the expected size (1,153 bp) in four of the six clones (b1,b2,c2,c3), suggesting that these clones had undergone disruption of the pbmap-2 locus. (C) The parental population P (which was used for the limiting dilution), clone a1 (with a wild-type Pbmap-2 locus) and the knockout clones b1 and c2 were selected for Southern blot analysis. DNA was digested with PacI or ClaI and probed with a fragment (66–727 bp) underlined in black in the targeting construct. The expected sizes of the restriction fragments are indicated in (A). The ClaI blot for the parental population P had bands representing the episomal (6.6 kb), wild-type and disrupted pbmap-2 loci. Both blots showed that clones b1 and c2 had a disrupted pbmap-2 locus. (D) RT–PCR with 1–1.5 μg of RNA from a1, b1 and c2 clones was performed with primers marked with a ‘^' in (A); primers 3,4 in supplementary Table 4 online). The region between the primers can be amplified only if full-length pbmap-2 is transcribed. In clone a1, we observed an amplification product of the expected size (869 bp) but not in clone b1 or c2 (lane 1 for each clone). We amplified LDH (primers 5,6) and pbs48 (primers 7,8; lanes 2 and 3, respectively, for each clone) as controls. No products were amplified from LDH reactions without reverse transcriptase (RT−).