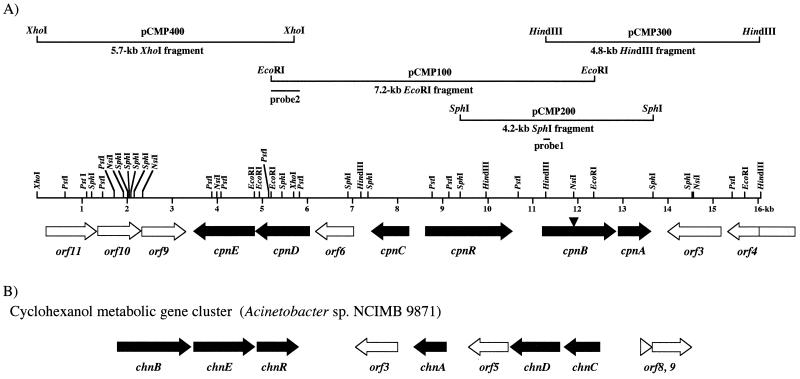
FIG. 2.
(A) Physical map of the cpn gene cluster in Comamonas sp. strain NCIMB 9872 and subclones in the 16,058-bp region. The orientation of the arrows indicates the direction of gene transcription. The black arrows are genes involved in cyclopentanol degradation. cpnR is a putative regulatory gene. Other ORFs are marked by open arrows. An arrowhead in the cpnB gene indicates the point of insertion of a lacZ-Kmr cassette at the NsiI site resulting in a mutant strain designated 9872MB. Probes 1 and 2 are PCR or restriction fragments used in hybridization experiments. (B) Gene organization of the cyclohexanol degradation (chn) pathway in Acinetobacter sp. strain 9871 (33). The chnABCDE genes (black arrows) encode cyclohexanol dehydrogenase, CHMO, caprolactone hydrolase, 6-hydroxyhexanoate dehydrogenase, and 6-oxohexanoate dehydrogenase, respectively; chnR is an AraC/XylS-type regulator (32, 33); orf5 is an equivalent of orf6 in the cpn pathway; orf3 encodes a potential pilin gene inverting protein; orf8 and -9 are putative transposases A and B, respectively (33). The chn gene cluster as shown is different from that present in strain SE19 (19) with the absence of “chnZ” in between chnR and orf3 (chnY designation in 19), and there are no equivalents of orf8 and -9 in the latter strain.