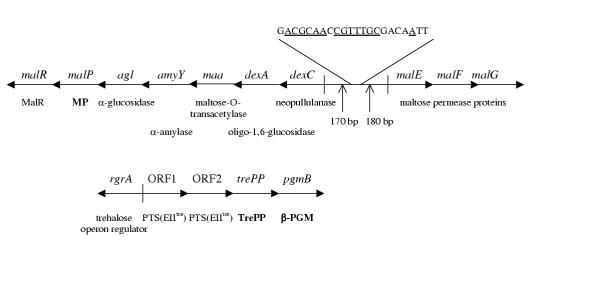
Figure 1.
The predicted maltose (above) and trehalose operons (below) of L. lactis. Genes/open reading frames are presented by arrows indicating the direction of transcription. Notations of enzymes of confirmed identity are shown in bold text. Other enzymes are from predictions based on amino acid sequence similarities. The DNA sequence shows the putative MalR binding site 170 bp and 180 bp from the translation start site of dexC and malE, respectively. The bases underlined are those that are identical to the bases found in the motifs of more than 50% of the 7 MalR binding sequences obtained from Schlösser et al. [17] and included in the current comparison. MalR, maltose operon regulator; MP, maltose phosphorylase; PTS(EIItre), trehalose-specific enzyme II component of the phosphotransferase system; TrePP, trehalose 6-phosphate phosphorylase; β-PGM, β-phosphoglucomutase. The figure is adapted from information on the genome sequence of L. lactis[5] and (GenBank accession no. Y18267) and from Nilsson and Rådström, Qian et al. and Andersson et al. [3,4,6].